from lec_utils import *
import matplotlib.pyplot as plt
Lecture 3¶
NumPy and Random Simulations¶
EECS 398: Practical Data Science, Spring 2025¶
practicaldsc.org • github.com/practicaldsc/sp25 • 📣 See latest announcements here on Ed
Agenda 📆¶
numpyarrays.- Multidimensional arrays and linear algebra.
- Randomness and simulations.
numpy arrays¶
Import statements¶
- We use
importstatements to add the objects (values, functions, classes) defined in other modules to our programs. There are a few different ways toimport.
Other terms I'll use for "module" are "library" and "package".
- Option 1:
import module.
Now, everytime we want to use a name in module, we must write module.<name>.
import math
math.sqrt(15)
3.872983346207417
sqrt(15)
--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[4], line 1 ----> 1 sqrt(15) NameError: name 'sqrt' is not defined
- Option 2:
import module as m.
Now, everytime we want to use a name in module, we can write m.<name> instead of module.<name>.
# This is the standard way that we will import numpy.
import numpy as np
np.pi
3.141592653589793
np.linalg.inv([[2, 1],
[3, 4]])
array([[ 0.8, -0.2],
[-0.6, 0.4]])
- Option 3:
from module import ....
This way, we explicitly state the names we want to import from module.
To import everything, write from module import *.
# Importing a particular function from the requests module.
from requests import get
# This typically fills up the namespace with a lot of unnecessary names, so use sparingly.
from math import *
sqrt
<function math.sqrt(x, /)>
NumPy¶

- NumPy (pronounced "num pie") is a Python library (module) that provides support for arrays and operations on them.
- The
pandaslibrary, which we will use for tabular data manipulation, works in conjunction withnumpy.
- To use
numpy, we need to import it. It's usually imported asnp(but doesn't have to be!)
We also had to install it on your computer first, but you already did that when you set up your environment.
import numpy as np
Arrays¶
- The core data structure in
numpyis the array. Moving forward, "array" will always refer to anumpyarray.
- One way to instantiate an array is to pass a list as an argument to the function
np.array.
np.array([4, 9, 1, 2])
array([4, 9, 1, 2])
- Arrays, unlike lists, must be homogenous – all elements must be of the same type.
# All elements are converted to strings!
np.array([1961, 'michigan'])
array(['1961', 'michigan'], dtype='<U21')
Array-number arithmetic¶
- Arrays make it easy to perform the same operation to every element without a
for-loop.
- This behavior is formally known as broadcasting. We also often say these operations are vectorized.
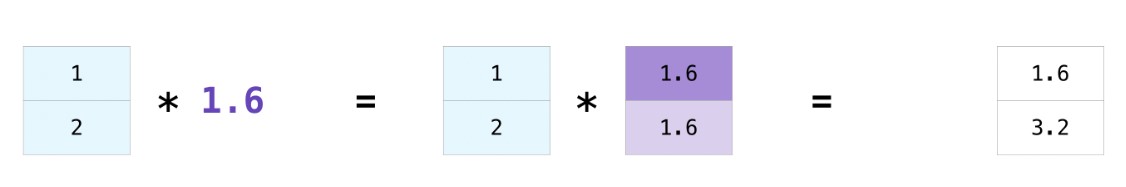
temps = [68, 72, 65, 64, 62, 61, 59, 64, 64, 63, 65, 62]
temps
[68, 72, 65, 64, 62, 61, 59, 64, 64, 63, 65, 62]
temp_array = np.array(temps)
# Increase all temperatures by 3 degrees.
temp_array + 3
array([71, 75, 68, 67, 65, 64, 62, 67, 67, 66, 68, 65])
# Halve all temperatures.
temp_array / 2
array([34. , 36. , 32.5, 32. , 31. , 30.5, 29.5, 32. , 32. , 31.5, 32.5,
31. ])
# Convert all temperatures to Celsius.
(5 / 9) * (temp_array - 32)
array([20. , 22.22, 18.33, 17.78, 16.67, 16.11, 15. , 17.78, 17.78,
17.22, 18.33, 16.67])
- Note: In none of the above cells did we actually modify
temp_array! Each of those expressions created a new array. To actually changetemp_array, we need to reassign it to a new array.
temp_array
array([68, 72, 65, 64, 62, 61, 59, 64, 64, 63, 65, 62])
temp_array = (5 / 9) * (temp_array - 32)
# Now in Celsius!
temp_array
array([20. , 22.22, 18.33, 17.78, 16.67, 16.11, 15. , 17.78, 17.78,
17.22, 18.33, 16.67])
⚠️ The dangers of unnecessary for-loops¶
- Under the hood,
numpyis implemented in C and Fortran, which are compiled languages that are much faster than Python. As a result, these vectorized operations are much quicker than if we used a vanilla Pythonfor-loop.
Also, the fact that arrays must be homogenous lend themselves to more efficient representations in memory.
- We can time code in a Jupyter Notebook. Let's try and square a long sequence of integers and see how long it takes with a Python loop:
%%timeit
squares = []
for i in range(1_000_000):
squares.append(i * i)
29.3 ms ± 82.1 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
- In vanilla Python, this takes about 0.03 seconds per loop.
Innumpy:
%%timeit
squares = np.arange(1_000_000) ** 2
866 μs ± 50.1 μs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
- Only takes about 0.0008 seconds per loop, approximately 40x faster!
Element-wise arithmetic¶
- We can apply arithmetic operations to multiple arrays, provided they have the same length.
- The result is computed element-wise, which means that the arithmetic operation is applied to one pair of elements from each array at a time.
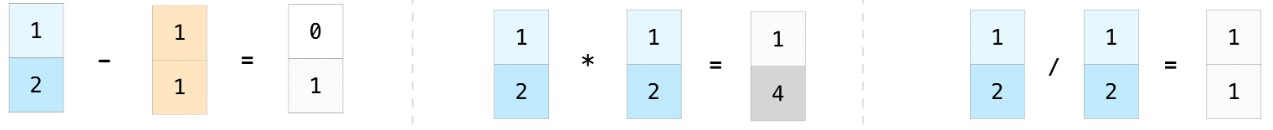
a = np.array([4, 5, -1])
b = np.array([2, 3, 2])
a + b
array([6, 8, 1])
a / b
array([ 2. , 1.67, -0.5 ])
a ** 2 + b ** 2
array([20, 34, 5])
# Broadcasting implicitly replaces the 3 with np.array([3, 3, 3]),
# so that the shapes match up.
a + 3
array([7, 8, 2])
Array methods¶
- Arrays come equipped with several handy methods; some examples are below, but you can read about them all here.
arr = np.array([3, 8, 4, -3.2])
(2 ** arr).sum()
280.108818820412
(2 ** arr).mean()
70.027204705103
(2 ** arr).max()
256.0
(2 ** arr).argmax()
1
# An attribute, not a method.
arr.shape
(4,)
Question 🤔 (Answer at practicaldsc.org/q)
What questions do we have about arrays so far?
Activity
🎉 Congrats! 🎉 You won the lottery 💰. Here's how your payout works: on the first day of September, you are paid \$0.01. Every day thereafter, your pay doubles, so on the second day you're paid \$0.02, on the third day you're paid \$0.04, on the fourth day you're paid \$0.08, and so on.
September has 30 days.
Write a one-line expression that uses the numbers 2 and 30, along with the function np.arange and at least one array method, that computes the total amount in dollars you will be paid in September. No for-loops or list comprehensions allowed!
We have a 🎥 walkthrough video of this problem (also posted on the course website under "⏯️ videos" for Lecture 3), but don't watch it until you've tried it yourself!
(2 ** np.arange(30) / 100).sum()
10737418.23
Boolean filtering¶
- Comparisons with arrays yield Boolean arrays! These can be used to answer questions about the values in an array.
temp_array
array([20. , 22.22, 18.33, 17.78, 16.67, 16.11, 15. , 17.78, 17.78,
17.22, 18.33, 16.67])
temp_array >= 18
array([ True, True, True, False, False, False, False, False, False,
False, True, False])
- How many values are greater than or equal to 18?
(temp_array >= 18).sum()
4
np.count_nonzero(temp_array >= 18)
4
- What fraction of values are greater than or equal to 18?
(temp_array >= 18).mean()
0.3333333333333333
- Which values are greater than or equal to 18?
temp_array[temp_array >= 18]
array([20. , 22.22, 18.33, 18.33])
- Which values are between 18 and 20?
# Note the parentheses!
temp_array[(temp_array >= 18) & (temp_array <= 20)]
array([20. , 18.33, 18.33])
# WRONG!
temp_array[(temp_array >= 18) and (temp_array <= 20)]
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) Cell In[43], line 2 1 # WRONG! ----> 2 temp_array[(temp_array >= 18) and (temp_array <= 20)] ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
Note: & and | vs. and and or¶
- In Python, the standard symbols for "and" and "or" are, literally,
andandor.
if (5 > 3 and 'h' + 'i' == 'hi') or (-2 > 0):
print('success')
success
- But, when taking the element-wise and/or of two arrays, the standard operators don't work.
Instead, use the bitwise operators:&for "and",|for "or".
temp_array
array([20. , 22.22, 18.33, 17.78, 16.67, 16.11, 15. , 17.78, 17.78,
17.22, 18.33, 16.67])
# Don't forget parentheses when using multiple conditions!
temp_array[(temp_array % 2 == 0) | (temp_array == temp_array.min())]
array([20., 15.])
- Read more about these differences here.
Multidimensional arrays and linear algebra¶
Multidimensional arrays¶
- A matrix can be represented in code using a two dimensional (2D) array.
- 2D arrays also resemble tables, or DataFrames, so it's worthwhile to study how they work.
nums = np.array([
[5, 1, 9, 7],
[9, 8, 2, 3],
[2, 5, 0, 4]
])
nums
array([[5, 1, 9, 7],
[9, 8, 2, 3],
[2, 5, 0, 4]])
# nums has 3 rows and 4 columns.
nums.shape
(3, 4)
- In addition to creating 2D arrays from scratch, we can also create 2D arrays by reshaping other arrays.
# Here, we're asking to reshape np.arange(1, 7)
# so that it has 2 rows and 3 columns.
a = np.arange(1, 7).reshape((2, 3))
a
array([[1, 2, 3],
[4, 5, 6]])
Operations along axes¶
- In 2D arrays (and DataFrames), axis 0 refers to the rows (up and down) and axis 1 refers to the columns (left and right).
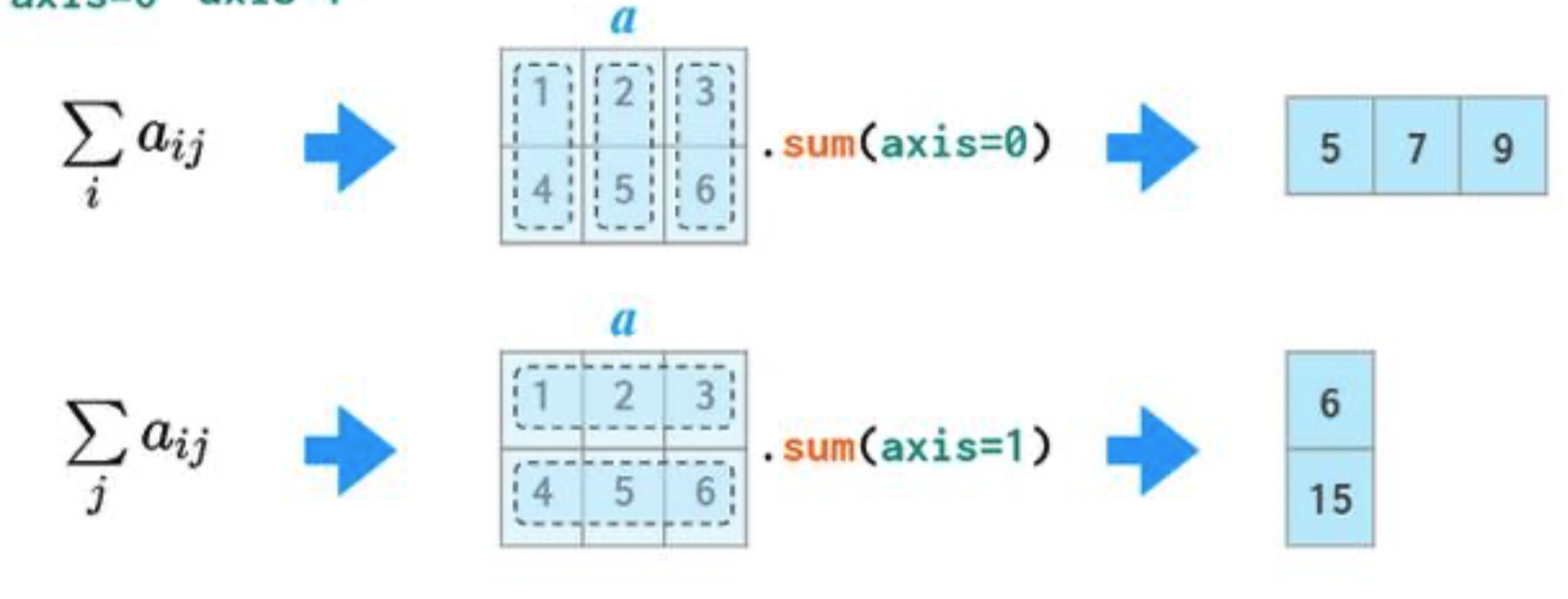
a
array([[1, 2, 3],
[4, 5, 6]])
- If we specify
axis=0,a.sumwill "compress" along axis 0.
a.sum(axis=0)
array([5, 7, 9])
- If we specify
axis=1,a.sumwill "compress" along axis 1.
a.sum(axis=1)
array([ 6, 15])
Selecting rows and columns from 2D arrays¶
- You can use
[square brackets]to slice rows and columns out of an array, too.
- The general convention is:
array[<row positions>, <column positions>]
a
array([[1, 2, 3],
[4, 5, 6]])
# Accesses row 0 and all columns.
a[0, :]
array([1, 2, 3])
# Same as the above.
a[0]
array([1, 2, 3])
# Accesses all rows and column 1.
a[:, 1]
array([2, 5])
# Access all rows and columns 0 and 2.
a[:, [0, 2]]
array([[1, 3],
[4, 6]])
# Accesses row 0 and columns 1 and onwards.
a[0, 1:]
array([2, 3])
Activity
Suppose we run the cell below.s = (5, 3)
grid = np.ones(s) * 2 * np.arange(1, 16).reshape(s)
grid[-1, 1:].sum()
What is the output of the cell? Try and answer without writing any code. See the annotated slides for the solution.
Example: Image processing¶
- Images can be represented as 3D
numpyarrays.
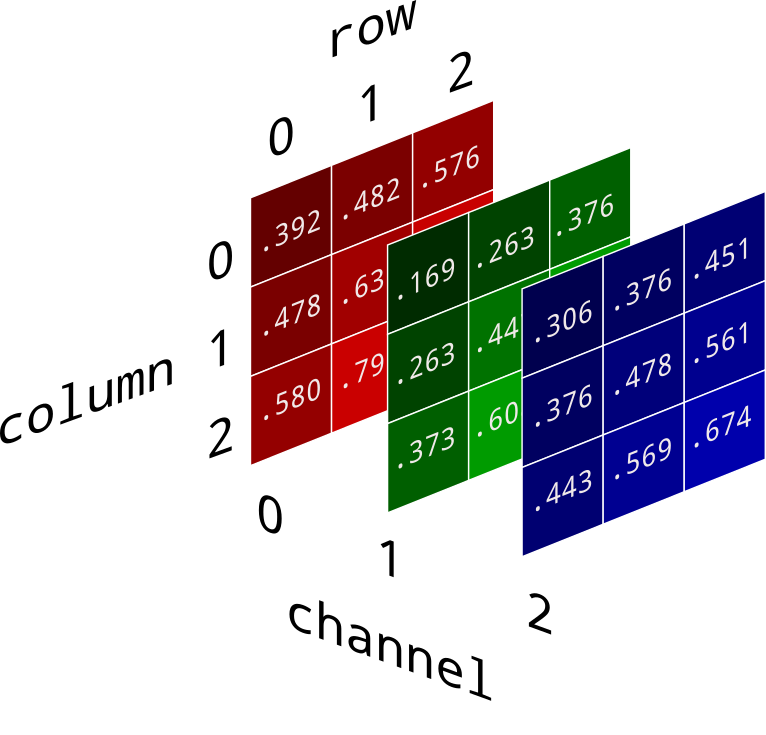 (image source)
(image source)- The color of each pixel can be described with three numbers under the RGB model – a red value, green value, and blue value. Each of these can vary from 0 (least intensity) to 255 (most intensity).
Experiment with RGB colors here.
img = np.asarray(Image.open('imgs/junior.jpeg'))
img
array([[[ 98, 62, 46],
[ 89, 56, 39],
[ 88, 56, 41],
...,
[123, 78, 59],
[125, 78, 60],
[128, 81, 63]],
[[ 96, 60, 44],
[ 89, 56, 39],
[ 89, 57, 42],
...,
[124, 79, 60],
[125, 78, 60],
[127, 80, 62]],
[[ 94, 58, 42],
[ 89, 56, 39],
[ 89, 57, 42],
...,
[125, 78, 60],
[125, 78, 60],
[126, 79, 61]],
...,
[[ 89, 50, 11],
[ 85, 46, 7],
[ 81, 42, 3],
...,
[ 94, 68, 33],
[100, 74, 39],
[108, 82, 47]],
[[ 86, 48, 11],
[ 84, 46, 9],
[ 84, 46, 9],
...,
[ 92, 66, 31],
[100, 74, 39],
[111, 85, 50]],
[[ 94, 56, 19],
[ 93, 55, 18],
[ 95, 57, 21],
...,
[ 90, 64, 29],
[ 98, 72, 37],
[112, 86, 51]]], dtype=uint8)
img.shape
(2048, 1536, 3)
plt.imshow(img)
plt.axis('off');
Applying a grayscale filter¶
- One way to convert an image to grayscale is to average its red, green, and blue values.
mean_2d = img.mean(axis=2)
mean_2d
array([[68.67, 61.33, 61.67, ..., 86.67, 87.67, 90.67],
[66.67, 61.33, 62.67, ..., 87.67, 87.67, 89.67],
[64.67, 61.33, 62.67, ..., 87.67, 87.67, 88.67],
...,
[50. , 46. , 42. , ..., 65. , 71. , 79. ],
[48.33, 46.33, 46.33, ..., 63. , 71. , 82. ],
[56.33, 55.33, 57.67, ..., 61. , 69. , 83. ]])
# This is just a single red channel!
plt.imshow(mean_2d.astype(int))
plt.axis('off');
- We need to repeat
mean_2dthree times along axis 2, to use the same values for the red, green, and blue channels.np.repeatwill help us here.
# np.newaxis is an alias for None.
# It helps us introduce an additional axis.
np.arange(5)[:, np.newaxis]
array([[0],
[1],
[2],
[3],
[4]])
np.repeat(np.arange(5)[:, np.newaxis], 3, axis=1)
array([[0, 0, 0],
[1, 1, 1],
[2, 2, 2],
[3, 3, 3],
[4, 4, 4]])
mean_3d = np.repeat(mean_2d[:, :, np.newaxis], 3, axis=2).astype(int)
mean_3d
array([[[68, 68, 68],
[61, 61, 61],
[61, 61, 61],
...,
[86, 86, 86],
[87, 87, 87],
[90, 90, 90]],
[[66, 66, 66],
[61, 61, 61],
[62, 62, 62],
...,
[87, 87, 87],
[87, 87, 87],
[89, 89, 89]],
[[64, 64, 64],
[61, 61, 61],
[62, 62, 62],
...,
[87, 87, 87],
[87, 87, 87],
[88, 88, 88]],
...,
[[50, 50, 50],
[46, 46, 46],
[42, 42, 42],
...,
[65, 65, 65],
[71, 71, 71],
[79, 79, 79]],
[[48, 48, 48],
[46, 46, 46],
[46, 46, 46],
...,
[63, 63, 63],
[71, 71, 71],
[82, 82, 82]],
[[56, 56, 56],
[55, 55, 55],
[57, 57, 57],
...,
[61, 61, 61],
[69, 69, 69],
[83, 83, 83]]])
plt.imshow(mean_3d)
plt.axis('off');
- Let's sepia-fy Junior!
 (Image credits)
(Image credits)
- From here, we can apply this conversion to each pixel.
sepia_filter = np.array([
[0.393, 0.769, 0.189],
[0.349, 0.686, 0.168],
[0.272, 0.534, 0.131]
])
# Multiplies each pixel by the sepia_filter matrix.
# Then, clips each RGB value to be between 0 and 1.
filtered = (img @ sepia_filter.T).clip(0, 255).astype(int)
filtered
array([[[ 94, 84, 65],
[ 85, 76, 59],
[ 85, 76, 59],
...,
[119, 106, 82],
[120, 107, 83],
[124, 110, 86]],
[[ 92, 82, 63],
[ 85, 76, 59],
[ 86, 77, 60],
...,
[120, 107, 83],
[120, 107, 83],
[123, 109, 85]],
[[ 89, 79, 62],
[ 85, 76, 59],
[ 86, 77, 60],
...,
[120, 107, 83],
[120, 107, 83],
[121, 108, 84]],
...,
[[ 75, 67, 52],
[ 70, 62, 48],
[ 64, 57, 44],
...,
[ 95, 84, 66],
[103, 92, 71],
[114, 101, 79]],
[[ 72, 64, 50],
[ 70, 62, 48],
[ 70, 62, 48],
...,
[ 92, 82, 64],
[103, 92, 71],
[118, 105, 82]],
[[ 83, 74, 57],
[ 82, 73, 57],
[ 85, 75, 59],
...,
[ 90, 80, 62],
[100, 89, 69],
[119, 106, 83]]])
plt.imshow(filtered)
plt.axis('off');
Matrix multiplication¶
- In the coming weeks, we'll start to rely more and more on tools from linear algebra.
You'll need this in Homework 2!
- Suppose the matrix $A$ and vectors $\vec x$ and $\vec y$ are defined as follows:
A = np.array([[2, -5, 1],
[0, 3, 2]])
x = np.array([[1],
[-1],
[4]])
y = np.array([[3],
[-2]])
- We can use
numpyto compute various quantities involving $A$, $\vec x$, and $\vec y$.
For instance, what is the result of the product $A \vec x$?
See the annotated slides for the math worked out.
A @ x
array([[11],
[ 5]])
- What is the result of the product $A^T \vec y$?
A.T @ y
array([[ 6],
[-21],
[ -1]])
- What is the result of the product $\vec x^T A^T A \vec x$?
x.T @ A.T @ A @ x
array([[146]])
Example: Fibonacci numbers¶
The sequence of Fibonacci numbers,
$$1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, ...$$
can be computed using matrix multiplication!
- It can be shown (with induction!) that if $f_1 = 1$, $f_2 = 1$, and $f_{n} = f_{n - 1} + f_{n - 2}, n \geq 2$, then:
fib = np.array([[1, 1],
[1, 0]])
np.linalg.matrix_power(fib, 8)
array([[34, 21],
[21, 13]])
Key takeaway: avoid for-loops whenever possible!¶
You can do a lot without for-loops, both in numpy and in pandas.
Randomness and simulations¶
np.random¶
- The submodule
np.randomcontains various functions that produce random results.
These use pseudo-random number generators to generate random-seeming sequences of results.
np.random.randomreturns a real number drawn at random from the interval [0, 1].
np.random.random()
0.6599195323554048
np.random.randintreturns a random integer in the specified range.
In the example below, between 1 and 6, inclusive.
# Run this cell multiple times!
np.random.randint(1, 7)
5
Random sampling¶
np.random.choiceandnp.random.multinomialallow us to draw random samples.
np.random.choicereturns randomly selected element(s) from the provided sequence.
By default, this is done with replacement, but it can be done without replacement, too.
unique_names = np.load('data/sp25-names.npy', allow_pickle=True)
unique_names
array(['Abhinav', 'Alvin', 'Andrew', ..., 'Toby', 'Trent', 'Yu'],
dtype=object)
# Returns a randomly selected element from the provided array, 5 times, with replacement.
# The resulting array COULD have duplicates.
np.random.choice(unique_names, 5)
array(['Hasan', 'Christian', 'Isaiah', 'Isaiah', 'Sean'], dtype=object)
# Returns a randomly selected element from the provided array, 5 times, without replacement.
# The resulting array CANNOT have duplicates.
np.random.choice(unique_names, 5, replace=False)
array(['Alvin', 'Yu', 'Abhinav', 'Joshua', 'Maxwell'], dtype=object)
np.random.multinomialreturns the result of drawing a sample from a multinomial distribution.
For instance, the cell below simulates the number of marbles of each color drawn from a bag in which 30% are blue, 50% are orange, and 20% are purple, and 15 marbles are drawn with replacement.
np.random.multinomial(15, [0.3, 0.5, 0.2])
array([4, 7, 4])
Simulations¶
- Often, we'll want to estimate the probability of an event, but it may not be possible – or we may not know how – to calculate the probability exactly.
e.g., the probability that I see between 40 and 50 heads when I flip a fair coin 100 times.
- Or, we may have a theoretical answer, and want to validate it using another approach.
- In such cases, we can use the power of simulation.
We can repeat the experiment many, many times, compute the fraction of experiments in which our event occurs, and use this fraction as an estimate of the probability of our event.
This is the basis of Monte Carlo Methods.
- Theory tells us that the more repetitions we perform of our experiment, the closer our fraction will be to the true probability of the event!
Specifically, the Law of Large Numbers tells us this.
Example: Coin flipping¶
- Question: What is the probability that I see between 40 and 50 heads, inclusive, when I flip a fair coin 100 times?
- To estimate this probability, we need to:
- Flip 100 fair coins and write down the number of heads,
- and repeat that process many, many times.
np.random.multinomial(100, [0.5, 0.5])
array([51, 49])
np.random.multinomial(100, [0.5, 0.5], 100_000)
array([[51, 49],
[49, 51],
[55, 45],
...,
[35, 65],
[49, 51],
[53, 47]])
# outcomes is an array with 100,000 elements,
# each of which is the number of heads in 100 simulated flips of a fair coin.
outcomes = np.random.multinomial(100, [0.5, 0.5], 100_000)[:, 0]
outcomes
array([59, 50, 49, ..., 50, 57, 52])
Estimating a probability from empirical results¶
px.histogram(outcomes, title='Number of Heads in 100 Simulated Flips of a Fair Coin')
- Our estimated probability of seeing between 40 and 50 heads is the fraction (proportion) of experiments in which we saw between 40 and 50 heads:
((outcomes >= 40) & (outcomes <= 50)).mean()
0.52015
- This is remarkably close to the true, theoretical answer, which can be computed using the binomial distribution.
from scipy.special import comb
sum([comb(100, k) for k in range(40, 51)]) / (2 ** 100)
0.5221945185847369
Question 🤔 (Answer at practicaldsc.org/q)
What questions do you have about our coin flipping simulation?
Example: Airplane seats ✈️¶
- A permutation of a sequence is a reshuffling of its elements.
np.random.permutationreturns a permutation of the specified sequence.
np.random.permutation(['A', 'B', 'C'])
array(['A', 'C', 'B'], dtype='<U1')
- Suppose a flight on Wolverine Airlines is scheduled for $n$ passengers, all of whom have an assigned seat.
- The airline loses track of seat assignments and everyone sits in a random seat.
What is the probability that nobody is in their originally assigned seat?
Simulating airplane seats¶
- Let's first simulate one hypothetical "plane".
def simulate_one_plane(n, display=False):
"""Simulates one plane of n people.
Returns True if nobody is in their originally assigned seat,
or False if at least one person is.
"""
poss = np.arange(1, n + 1)
shuffled = np.random.permutation(poss)
if display:
print('Real plane: ', poss)
print('Simulated plane:', shuffled)
return (poss != shuffled).all()
simulate_one_plane(10, display=True)
Real plane: [ 1 2 3 4 5 6 7 8 9 10] Simulated plane: [ 4 10 7 5 8 1 3 9 6 2]
True
- Now, let's call
simulate_one_planemany, many times and compute the proportion of calls that returnTrue.
We'll arbitrarily start with $n = 50$, but change it and see what happens to the answer!
n = 50
prob = np.mean([simulate_one_plane(n) for _ in range(100_000)])
prob
0.36796
Airplane seats, solved¶
- The probability that nobody is in their originally assigned seat is:
- Fact: As $n \rightarrow \infty$, this approaches:
1 / np.e
0.36787944117144233
Walkthrough video of airplane seats example¶
- If we don't finish the airplane seats example in class, there's a walkthrough video of it below.
The walkthrough can also be found directly on the course website, by clicking the "⏯️ videos" button for Lecture 3.
YouTubeVideo('shc1YcPrfXE')
Curious to learn more?¶
- Watch the following video by Numberphile, a popular YouTube channel for math-related content.
YouTubeVideo('pbXg5EI5t4c')
- Simulations are powerful!