from lec_utils import *
def sample_from_pop(n=100):
x = np.linspace(-2, 3, n)
y = x ** 3 + (np.random.normal(0, 3, size=n))
return pd.DataFrame({'x': x, 'y': y})
sample_1 = sample_from_pop()
Announcements 📣¶
- Homework 8 is due on Friday (not today!).
- Homework 7 solutions are available at #259 on Ed.
- Check out the new FAQs page on the course website.
It has answers to frequently-asked theoretical questions. - The IA application is out for next semester and is due on Monday! See #238 on Ed for more details.
Agenda¶
- Recap: Multiple linear regression and feature engineering.
- Numerical-to-numerical transformations.
- The modeling recipe, revisited.
OneHotEncoderand multicollinearity.StandardScalerand standardized regression coefficients.
Question 🤔 (Answer at practicaldsc.org/q)
Remember that you can always ask questions anonymously at the link above!
Recap: Multiple linear regression and feature engineering¶
The general problem¶
- We have $n$ data points, $\left({ \vec x_1}, {y_1}\right), \left({ \vec x_2}, {y_2}\right), \ldots, \left({ \vec x_n}, {y_n}\right)$,
where each $ \vec x_i$ is a feature vector of $d$ features: $${\vec{x_i}} = \begin{bmatrix} {x^{(1)}_i} \\ {x^{(2)}_i} \\ \vdots \\ {x^{(d)}_i} \end{bmatrix}$$
- We want to find a good linear hypothesis function:
The general solution¶
- Define the design matrix $ X \in \mathbb{R}^{n \times (d + 1)}$ and observation vector $ \vec y \in \mathbb{R}^n$:
Then, solve the normal equations to find the optimal parameter vector, $\vec{w}^*$:
$${ X^TX} \vec{w}^* = { X^T} { \vec y}$$
The goal of feature engineering¶
- Feature engineering is the act of finding transformations that transform data into effective quantitative variables.
Put simply: feature engineering is creating new features using existing features.
- Example: One hot encoding.
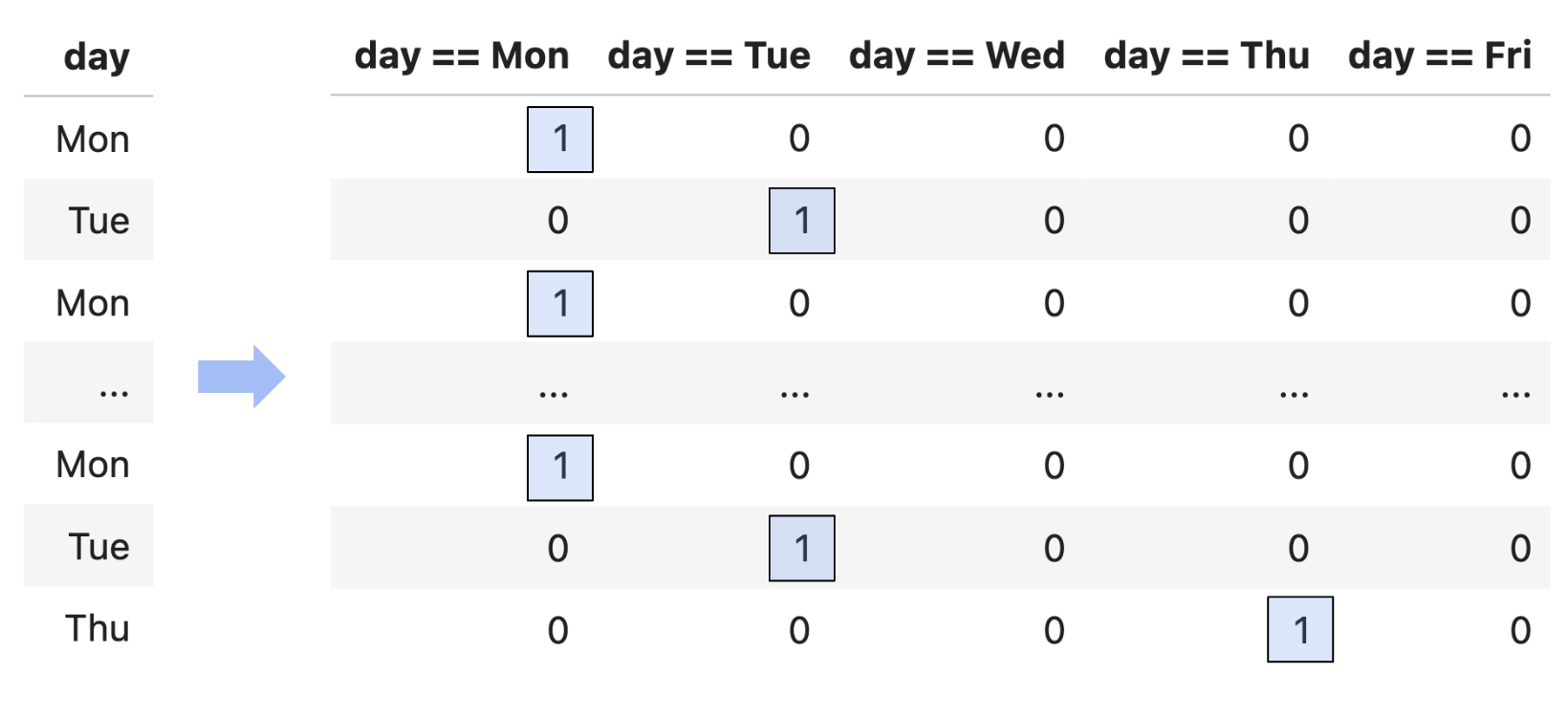
- Example: Numerical-to-numerical transformations.
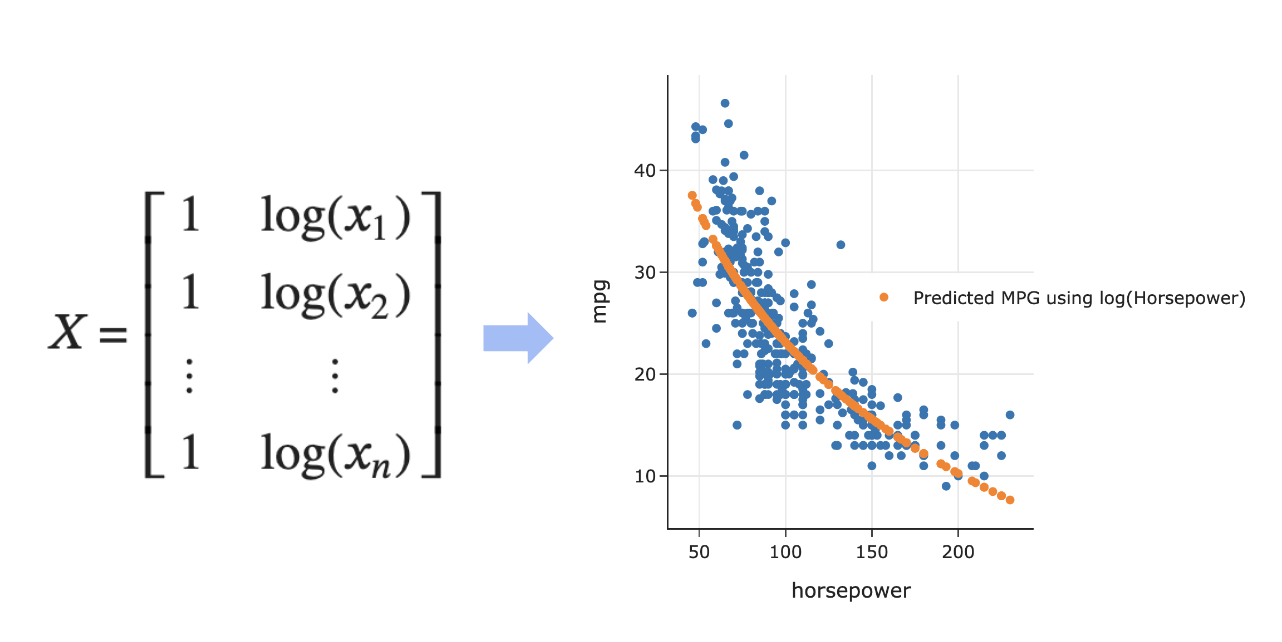
Numerical-to-numerical transformations¶
Linearization¶
- The Tukey Mosteller Bulge Diagram helps us pick which numerical-to-numerical transformations to apply to data in order to linearize it.
Alternative interpretation: it helps us determine which features to create.
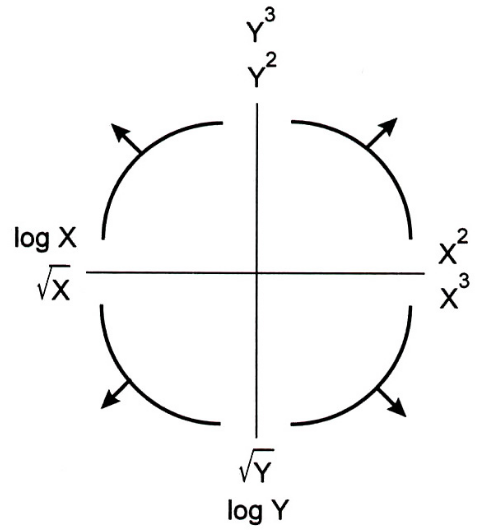
- Why? We're working with linear models. The more linear our data looks in terms of its features, the better we'll able to model the data.
Example: Polynomial regression¶
- Last class, we engineered a new feature by taking the $\log$ of an existing feature; this is a numerical-to-numerical transformation.
This allowed us to create a curve of best fit.
- Consider the dataset below.
sample_1is defined at the top of this notebook.
px.scatter(sample_1, x='x', y='y')
- A simple linear regression line isn't sufficient enough to model the relationship between the two variables.
Example: Polynomial regression¶
- The scatter plot appears to roughly resemble a degree 3 (cubic) polynomial, so let's try and fit a degree 3 polynomial. This will involve creating a design matrix with quadratic and cubic features.
X = sample_1[['x']].copy()
X
| x | |
|---|---|
| 0 | -2.00 |
| 1 | -1.95 |
| 2 | -1.90 |
| ... | ... |
| 97 | 2.90 |
| 98 | 2.95 |
| 99 | 3.00 |
100 rows × 1 columns
# Note that X itself is not the design matrix;
# sklearn's LinearRegression object will create the needed design matrix
# by adding a column of 1s to the start of X.
X['x^2'] = X['x'] ** 2
X['x^3'] = X['x'] ** 3
X
| x | x^2 | x^3 | |
|---|---|---|---|
| 0 | -2.00 | 4.00 | -8.00 |
| 1 | -1.95 | 3.80 | -7.41 |
| 2 | -1.90 | 3.61 | -6.85 |
| ... | ... | ... | ... |
| 97 | 2.90 | 8.40 | 24.36 |
| 98 | 2.95 | 8.70 | 25.66 |
| 99 | 3.00 | 9.00 | 27.00 |
100 rows × 3 columns
- Now, let's fit a
LinearRegressionmodel fromsklearnand look at the resulting predictions.
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X=X, y=sample_1['y'])
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
model.predict(X)
array([-9.72, -9.03, -8.38, ..., 24.1 , 25.47, 26.89])
fig = px.scatter(sample_1, x='x', y='y')
fig.add_trace(go.Scatter(
x=X['x'],
y=model.predict(X),
mode='lines',
line=dict(width=5),
name='Degree 3 Polynomial of Best Fit'
))
- The orange curve above is of the form:
model.intercept_
-0.5198355347237897
model.coef_
array([-0.31, -0.21, 1.12])
- While the curve is non-linear, it is linear in the parameters.
- Amdahl's Law relates the runtime of a program on $p$ processors to the time to do the sequential and nonsequential parts on one processor.
- Collect data by timing a program with varying numbers of processors:
| Processors | Time (Hours) |
|---|---|
| 1 | 8 |
| 2 | 4 |
| 4 | 3 |
- To find $w_0^*$ and $w_1^*$ in the hypothesis function $H(x) = w_0 + w_1 \cdot \frac{1}{x}$, we need to create an appropriate design matrix:
- Then, the problem reduces to finding the parameter vector, $\vec{w}^*$, that solves the normal equations, $(X^TX)^{-1}X^T \vec{y}$.
Question 🤔 (Answer at practicaldsc.org/q)
Which hypothesis function is not linear in the parameters?
- A. $H(\vec{x}) = w_1 (x^{(1)} x^{(2)}) + \frac{w_2}{x^{(1)}} \sin \left( x^{(2)} \right)$
- B. $H(\vec{x}) = 2^{w_1} x^{(1)}$
- C. $H(\vec{x}) = \vec{w} \cdot \text{Aug}(\vec{x})$
- D. $H(\vec{x}) = w_1 \cos (x^{(1)}) + w_2 2^{x^{(2)} \log x^{(3)}}$
- E. More than one of the above.
How do we fit hypothesis functions that aren't linear in the parameters?¶
- Suppose we want to fit the hypothesis function:
- This is not linear in terms of $w_0$ and $w_1$, so our results for linear regression don't apply.
- Possible solution: Try to transform the above equation so that it is linear in some other parameters, by applying an operation to both sides.
- Suppose we take the $\log$ of both sides of the equation.
- Then, using properties of logarithms, we have:
- Solution: Create a new hypothesis function, $T(x)$, with parameters $b_0$ and $b_1$, where $T(x) = b_0 + b_1 x$.
- This hypothesis function is related to $H(x)$ by the relationship $T(x) = \log H(x)$.
- $\vec{b}$ is related to $\vec{w}$ by $b_0 = \log w_0$ and $b_1 = w_1$.
- Our new observation vector, $\vec{z}$, is $\begin{bmatrix} \log y_1 \\ \log y_2 \\ ... \\ \log y_n \end{bmatrix}$.
- $T(x) = b_0 + b_1x$ is linear in its parameters, $b_0$ and $b_1$.
- Use the solution to the normal equations to find $\vec{b}^*$, and the relationship between $\vec{b}$ and $\vec{w}$ to find $\vec{w}^*$.
The modeling recipe, revisited¶
The original modeling recipe, from Lecture 14¶
- Choose a model.
- Choose a loss function.
- Minimize average loss to find optimal model parameters.
The updated modeling recipe¶
- Create, or engineer, features to best reflect the "meaning" behind data.
Recently, we've done this with one hot encoding and numerical-to-numerical transformations.
- Choose a model.
Recently, we've used the simple/multiple linear regression model.
- Choose a loss function.
Recently, we've mostly used squared loss.
- Minimize average loss (empirical risk) to find optimal model parameters, $\vec{w}^*$.
Originally, we had to use calculus or linear algebra to minimize empirical risk, but more recently we've just usedmodel.fit. This step is also called fitting the model to the data.
- Evaluate the performance of the model in relation to other models, using MSE or $R^2$.
- We can do all of the above directly in
sklearn!

preprocessing and linear_models¶
- For the feature engineering step of the modeling pipeline, we will use
sklearn'spreprocessingmodule.
- For the model creation step of the modeling pipeline, we will use
sklearn'slinear_modelmodule, as we've already seen.linear_model.LinearRegressionis an example of an estimator class.
Transformer classes¶
- Transformers take in "raw" data and output "processed" data. They are used for creating features.
- Transformers, like most relevant features of
sklearn, are classes, not functions, meaning you need to instantiate them and call their methods.
- Today, we'll introduce two transformer classes. We'll look at how to write code to use each one, but also discuss some of the underlying statistical nuances.
- Next class, we'll see how to chain transformers and estimators together into larger Pipelines.
OneHotEncoder and multicollinearity¶
Example: Commute times 🚗¶
- For this first example, we'll continue working with our commute times dataset.
df = pd.read_csv('data/commute-times.csv')
df['day_of_month'] = pd.to_datetime(df['date']).dt.day
df['month'] = pd.to_datetime(df['date']).dt.month_name()
df.head()
| date | day | home_departure_time | home_departure_mileage | ... | work_departure_time_hr | mileage_to_home | day_of_month | month | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 5/15/2023 | Mon | 2023-05-15 10:49:00 | 15873.0 | ... | 17.17 | 53.0 | 15 | May |
| 1 | 5/16/2023 | Tue | 2023-05-16 07:45:00 | 15979.0 | ... | NaN | NaN | 16 | May |
| 2 | 5/22/2023 | Mon | 2023-05-22 08:27:00 | 50407.0 | ... | 15.90 | 54.0 | 22 | May |
| 3 | 5/23/2023 | Tue | 2023-05-23 07:08:00 | 50535.0 | ... | NaN | NaN | 23 | May |
| 4 | 5/30/2023 | Tue | 2023-05-30 09:09:00 | 50664.0 | ... | 17.12 | 54.0 | 30 | May |
5 rows × 20 columns
- We'll focus specifically on the
'day'and'month'columns.
df[['day', 'month']]
| day | month | |
|---|---|---|
| 0 | Mon | May |
| 1 | Tue | May |
| 2 | Mon | May |
| ... | ... | ... |
| 62 | Mon | March |
| 63 | Tue | March |
| 64 | Thu | March |
65 rows × 2 columns
Example transformer: OneHotEncoder¶
- Last class, we had to manually one hot encode the
'day'column. Let's figure out how to one hot encode it automatically, along with the new'month'column.
df[['day', 'month']]
| day | month | |
|---|---|---|
| 0 | Mon | May |
| 1 | Tue | May |
| 2 | Mon | May |
| ... | ... | ... |
| 62 | Mon | March |
| 63 | Tue | March |
| 64 | Thu | March |
65 rows × 2 columns
- First, we need to import the relevant class from
sklearn.preprocessing.
It's best practice to import just the relevant classes you need fromsklearn.
from sklearn.preprocessing import OneHotEncoder
- Like an estimator, we need to instantiate and fit our
OneHotEncoderinstsance before it can transform anything.
ohe = OneHotEncoder()
# Error!
ohe.transform(df[['day', 'month']])
--------------------------------------------------------------------------- NotFittedError Traceback (most recent call last) Cell In[16], line 2 1 # Error! ----> 2 ohe.transform(df[['day', 'month']]) File ~/miniforge3/envs/pds/lib/python3.10/site-packages/sklearn/utils/_set_output.py:313, in _wrap_method_output.<locals>.wrapped(self, X, *args, **kwargs) 311 @wraps(f) 312 def wrapped(self, X, *args, **kwargs): --> 313 data_to_wrap = f(self, X, *args, **kwargs) 314 if isinstance(data_to_wrap, tuple): 315 # only wrap the first output for cross decomposition 316 return_tuple = ( 317 _wrap_data_with_container(method, data_to_wrap[0], X, self), 318 *data_to_wrap[1:], 319 ) File ~/miniforge3/envs/pds/lib/python3.10/site-packages/sklearn/preprocessing/_encoders.py:1008, in OneHotEncoder.transform(self, X) 985 def transform(self, X): 986 """ 987 Transform X using one-hot encoding. 988 (...) 1006 returned. 1007 """ -> 1008 check_is_fitted(self) 1009 transform_output = _get_output_config("transform", estimator=self)["dense"] 1010 if transform_output != "default" and self.sparse_output: File ~/miniforge3/envs/pds/lib/python3.10/site-packages/sklearn/utils/validation.py:1661, in check_is_fitted(estimator, attributes, msg, all_or_any) 1658 raise TypeError("%s is not an estimator instance." % (estimator)) 1660 if not _is_fitted(estimator, attributes, all_or_any): -> 1661 raise NotFittedError(msg % {"name": type(estimator).__name__}) NotFittedError: This OneHotEncoder instance is not fitted yet. Call 'fit' with appropriate arguments before using this estimator.
# Need to fit first.
ohe.fit(df[['day', 'month']])
OneHotEncoder()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
OneHotEncoder()
- Once we've fit, when we use the
transformmethod, we get a result we might not expect.
ohe.transform(df[['day', 'month']])
<65x16 sparse matrix of type '<class 'numpy.float64'>' with 130 stored elements in Compressed Sparse Row format>
- Since the resulting matrix is sparse – most of its elements are 0 –
sklearnuses a more efficient representation than a regularnumpyarray. We can convert to a regular (dense) array:
ohe.transform(df[['day', 'month']]).toarray()
array([[0., 1., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 1., 0., ..., 0., 0., 0.],
...,
[0., 1., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.]])
- The column names from
df[['day', 'month']]don't appear in the output above. We can use theget_feature_names_outmethod onoheto access the names and order of the one hot encoded columns, though:
ohe.get_feature_names_out()
array(['day_Fri', 'day_Mon', 'day_Thu', 'day_Tue', 'day_Wed',
'month_August', 'month_December', 'month_February',
'month_January', 'month_July', 'month_June', 'month_March',
'month_May', 'month_November', 'month_October', 'month_September'],
dtype=object)
pd.DataFrame(ohe.transform(df[['day', 'month']]).toarray(),
columns=ohe.get_feature_names_out()) # If we need a DataFrame back, for some reason.
| day_Fri | day_Mon | day_Thu | day_Tue | ... | month_May | month_November | month_October | month_September | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 1.0 | 0.0 | 0.0 | ... | 1.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 1.0 | 0.0 | 0.0 | 0.0 |
| 2 | 0.0 | 1.0 | 0.0 | 0.0 | ... | 1.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 62 | 0.0 | 1.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 |
| 63 | 0.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 |
| 64 | 0.0 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 |
65 rows × 16 columns
- Usually, we won't perform all of these intermediate steps, since the
OneHotEncoderwill be part of a larger Pipeline.
Example: Heights and weights¶
- We now know how to use
OneHotEncoder.
- To illustrate a mathematical issue involving one hot encoding, let's load in another dataset, this time containing the weights and heights of 25,000 18 year olds, taken from here.
people = pd.read_csv('data/heights-weights.csv').drop(columns=['Index'])
people.head()
| Height (Inches) | Weight (Pounds) | |
|---|---|---|
| 0 | 65.78 | 112.99 |
| 1 | 71.52 | 136.49 |
| 2 | 69.40 | 153.03 |
| 3 | 68.22 | 142.34 |
| 4 | 67.79 | 144.30 |
people.plot(kind='scatter', x='Height (Inches)', y='Weight (Pounds)',
title='Weight vs. Height for 25,000 18 Year Olds')
Motivating example¶
- Suppose we fit a simple linear regression model that uses height in inches to predict weight in pounds.
X = people[['Height (Inches)']]
y = people['Weight (Pounds)']
people_one_feat = LinearRegression()
people_one_feat.fit(X, y)
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
- $w_0^*$ and $w_1^*$ are shown below, along with the model's MSE on the data we used to train it.
We call this the model's training MSE.
people_one_feat.intercept_, people_one_feat.coef_
(-82.57574306454093, array([3.08]))
from sklearn.metrics import mean_squared_error
mean_squared_error(y, people_one_feat.predict(X))
101.58853248632849
An added feature¶
- Now, suppose we fit another regression model, that uses height in inches AND height in centimeters to predict weight.
people['Height (cm)'] = people['Height (Inches)'] * 2.54 # 1 inch = 2.54 cm.
X2 = people[['Height (Inches)', 'Height (cm)']]
people_two_feat = LinearRegression()
people_two_feat.fit(X2, y)
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
- What are $w_0^*$, $w_1^*$, $w_2^*$, and the model's MSE?
people_two_feat.intercept_, people_two_feat.coef_
(-82.57525639659859, array([ 3.38e+10, -1.33e+10]))
mean_squared_error(y, people_two_feat.predict(X2))
101.58853251653949
- Observation: The intercept is the same as before (roughly -82.57), as is the MSE. However, the coefficients on
'Height (Inches)'and'Height (cm)'are massive in size!
- It should be unsurprising that the MSE is the same, because the span of the design matrix is the same. So, the best predictions should be the same, too.
- But what's going on with the coefficients?
Redundant features¶
- Let's use simpler numbers for illustration. Suppose in the first model, $w_0^* = -80$ and $w_1^* = 3$.
- In the second model, we have:
- In the first model, we already found the "best" intercept ($-80$) and slope ($3$) in a linear model that uses height in inches to predict weight.
- So, as long as $w_1^* + 2.54 \cdot w_2^* = 3$ in the second model, the second model's predictions will be the same as the first, and hence they will also minimize MSE.
Infinitely many parameter choices¶
- Issue: There are an infinite number of $w_1^*$ and $w_2^*$ that satisfy $w_1^* + 2.54 \cdot w_2^* = 3$!
- Both hypothesis functions look very different, but actually make the same predictions.
model.coef_could return either set of coefficients, or any other of the infinitely many options.
- But neither set of coefficients is has any meaning!
(-80 - 10 * people.iloc[:, 0] + (13 / 2.54) * people.iloc[:, 2]).head()
0 117.35 1 134.55 2 128.20 3 124.65 4 123.36 dtype: float64
(-80 + 10 * people.iloc[:, 0] - (7 / 2.54) * people.iloc[:, 2]).head()
0 117.35 1 134.55 2 128.20 3 124.65 4 123.36 dtype: float64
Multicollinearity¶
- Multicollinearity occurs when features in a regression model are highly correlated with one another.
In other words, multicollinearity occurs when a feature can be predicted using a linear combination of other features, fairly accurately.
- When multicollinearity is present in the features, the coefficients in the model are uninterpretable – they have no meaning.
A "slope" represents "the rate of change of $y$ with respect to a feature", when all other features are held constant – but if there's multicollinearity, you can't hold other features constant.
- Note: Multicollinearity doesn't impact a model's predictions!
- It doesn't impact a model's ability to generalize to unseen data.
- If features are multicollinear in the data we've seen, they will probably be multicollinear in the data we haven't seen, drawn from the same distribution.
- Solutions:
- Manually remove highly correlated features.
- Use a dimensionality reduction technique (such as PCA) to automatically reduce dimensions.
One hot encoding and multicollinearity¶
- One hot encoding will result in multicollinearity unless you drop one of the one hot encoded features.
- Suppose we have the following fitted model:
For illustration, assume'weekend'was originally a categorical feature with two possible values,'Yes'or'No'.
- This is equivalent to:
- Note that for a particular row in the dataset, $\text{weekend==Yes} + \text{weekend==No}$ is always equal to 1.
- This means that the columns of the design matrix, $X$, for this model is not linearly independent, since the column of all 1s can be written as a linear combination of the $\text{weekend==Yes}$ and $\text{weekend==No}$ columns.
- This means that the design matrix is not full rank, which means that $X^TX$ is not invertible.
- This means that there are infinitely many possible solutions $\vec{w}^*$ to the normal equations, $(X^TX) \vec{w} = X^T\vec{y}$!
That's a problem, because we don't know which of these infinitely many solutionsmodel.coef_will find for us, and it's impossible to interpret the resulting coefficients, as we saw on the last slide.
- Solution: Drop one of the one hot encoded columns.
OneHotEncoderhas an option to do this.
OneHotEncoder returns¶
- Let's switch back to the commute times dataset,
df.
df[['day', 'month']]
| day | month | |
|---|---|---|
| 0 | Mon | May |
| 1 | Tue | May |
| 2 | Mon | May |
| ... | ... | ... |
| 62 | Mon | March |
| 63 | Tue | March |
| 64 | Thu | March |
65 rows × 2 columns
- Let's try using
drop='first'when instantiating aOneHotEncoder.
ohe_drop_one = OneHotEncoder(drop='first')
ohe_drop_one.fit(df[['day', 'month']])
OneHotEncoder(drop='first')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
OneHotEncoder(drop='first')
- How many features did the resulting transformer create?
len(ohe_drop_one.get_feature_names_out())
14
- Where did this number come from?
df['day'].nunique()
5
df['month'].nunique()
11
Key takeaways¶
- Multicollinearity is present in a linear model when one feature can be accurately predicted using one or more other features.
In other words, it is present when a feature is redundant.
- Multicollinearity doesn't pose an issue for prediction; it doesn't hinder a model's ability to generalize. Instead, it renders the coefficients of a linear model meaningless.
StandardScaler and standardized regression coefficients¶
Example: Predicting sales 📈¶
- To illustrate the next transformer class, we'll introduce a new dataset.
sales = pd.read_csv('data/sales.csv')
sales.head()
| net_sales | sq_ft | inventory | advertising | district_size | competing_stores | |
|---|---|---|---|---|---|---|
| 0 | 231.0 | 3.0 | 294 | 8.2 | 8.2 | 11 |
| 1 | 156.0 | 2.2 | 232 | 6.9 | 4.1 | 12 |
| 2 | 10.0 | 0.5 | 149 | 3.0 | 4.3 | 15 |
| 3 | 519.0 | 5.5 | 600 | 12.0 | 16.1 | 1 |
| 4 | 437.0 | 4.4 | 567 | 10.6 | 14.1 | 5 |
- For each of 26 stores, we have:
- net sales,
- square feet,
- inventory,
- advertising expenditure,
- district size, and
- number of competing stores.
- Our goal is to predict
'net_sales'as a function of other features.
An initial model¶
sales.head()
| net_sales | sq_ft | inventory | advertising | district_size | competing_stores | |
|---|---|---|---|---|---|---|
| 0 | 231.0 | 3.0 | 294 | 8.2 | 8.2 | 11 |
| 1 | 156.0 | 2.2 | 232 | 6.9 | 4.1 | 12 |
| 2 | 10.0 | 0.5 | 149 | 3.0 | 4.3 | 15 |
| 3 | 519.0 | 5.5 | 600 | 12.0 | 16.1 | 1 |
| 4 | 437.0 | 4.4 | 567 | 10.6 | 14.1 | 5 |
- No transformations are needed to predict
'net_sales'.
sales_model = LinearRegression()
sales_model.fit(X=sales.iloc[:, 1:], y=sales.iloc[:, 0])
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
- Suppose we're interested in learning how the various features impact
'net_sales', rather than just predicting'net_sales'for a new store. We'd then look at the coefficients.
sales_model.coef_
array([16.2 , 0.17, 11.53, 13.58, -5.31])
pd.DataFrame().assign(
column=sales.columns[1:],
original_coef=sales_model.coef_,
).set_index('column')
| original_coef | |
|---|---|
| column | |
| sq_ft | 16.20 |
| inventory | 0.17 |
| advertising | 11.53 |
| district_size | 13.58 |
| competing_stores | -5.31 |
- What do you notice?
sales.iloc[:, 1:]
| sq_ft | inventory | advertising | district_size | competing_stores | |
|---|---|---|---|---|---|
| 0 | 3.0 | 294 | 8.2 | 8.2 | 11 |
| 1 | 2.2 | 232 | 6.9 | 4.1 | 12 |
| 2 | 0.5 | 149 | 3.0 | 4.3 | 15 |
| ... | ... | ... | ... | ... | ... |
| 24 | 3.5 | 382 | 9.8 | 11.5 | 5 |
| 25 | 5.1 | 590 | 12.0 | 15.7 | 0 |
| 26 | 8.6 | 517 | 7.0 | 12.0 | 8 |
27 rows × 5 columns
Which features are most "important"?¶
- The most important feature is not necessarily the feature with largest magnitude coefficient, because different features may be on different scales.
- Suppose I fit two hypothesis functions:
- $H_1$ has store size measured in square feet.
- $H_2$ has store size measured in square meters.
- Store size is just as important in both hypothesis functions.
- But 1 square meter $\approx 10.76$ square feet, so the sizes in square meters will be 10.76x smaller.
- So, the coefficient of store size in $H_2$ will be 10.76 times larger than the coefficient of store size in $H_1$.
Intuition: if the values themselves are smaller, you need to multiply them by bigger coefficients to get the same predictions!
- Solution: If you care about the interpretability of the resulting coefficients, standardize each feature before performing regression.
Standardization¶
- Recall: to standardize a feature $x_1, x_2, ..., x_n$, we use the formula: $$z(x_i) = \frac{x_i - \bar{x}}{\sigma_x}$$
Example: 1, 7, 7, 9.
- Mean: $\frac{1 + 7 + 7 + 9}{4} = \frac{24}{4} = 6$.
- Standard deviation:
$$\text{SD} = \sqrt{\frac{1}{4} \left( (1-6)^2 + (7-6)^2 + (7-6)^2 + (9-6)^2 \right)} = \sqrt{\frac{1}{4} \cdot 36} = 3$$
- Standardized data:
$$1 \mapsto \frac{1-6}{3} = \boxed{-\frac{5}{3}} \qquad 7 \mapsto \frac{7-6}{3} = \boxed{\frac{1}{3}} \qquad 7 \mapsto \boxed{\frac{1}{3}} \qquad 9 \mapsto \frac{9-6}{3} = \boxed{1}$$
Example transformer: StandardScaler¶
StandardScalerstandardizes data using the mean and standard deviation of the data, as described on the previous slide.
- Like
OneHotEncoder,StandardScalerrequires some knowledge (mean and SD) of the dataset before transforming, so we need tofitanStandardScalertransformer before we can use thetransformmethod.
from sklearn.preprocessing import StandardScaler
stdscaler = StandardScaler()
# This is like saying "determine the mean and SD of each column in sales,
# other than the 'net_sales' column".
stdscaler.fit(sales.iloc[:, 1:])
StandardScaler()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
StandardScaler()
- Now, we can standardize any dataset, using the mean and standard deviation of the columns in
sales.iloc[:, 1:]. Typical usage is to fit transformer on a sample and use that already-fit transformer to transform future data.
stdscaler.transform([[5, 300, 10, 15, 6]])
/Users/surajrampure/miniforge3/envs/pds/lib/python3.10/site-packages/sklearn/base.py:493: UserWarning: X does not have valid feature names, but StandardScaler was fitted with feature names
array([[ 0.85, -0.47, 0.51, 1.05, -0.36]])
stdscaler.transform(sales.iloc[:, 1:].tail(5))
array([[-1.13, -1.31, -1.35, -1.6 , 0.89],
[ 0.14, 0.39, 0.4 , 0.32, -0.36],
[ 0.09, -0.03, 0.46, 0.36, -0.57],
[ 0.9 , 1.08, 1.05, 1.19, -1.61],
[ 2.67, 0.69, -0.3 , 0.46, 0.05]])
- We can peek under the hood and see what it computed!
stdscaler.mean_
array([ 3.33, 387.48, 8.1 , 9.69, 7.74])
stdscaler.var_
array([ 3.89, 35191.58, 13.72, 25.44, 23.08])
- If needed, the
fit_transformmethod will fit the transformer and then transform the data in one go.
new_scaler = StandardScaler()
new_scaler.fit_transform(sales.iloc[:, 1:].tail(5))
array([[-1.33, -1.79, -1.71, -1.88, 1.48],
[-0.32, 0.28, 0.43, 0.19, -0.05],
[-0.36, -0.24, 0.49, 0.23, -0.31],
[ 0.29, 1.11, 1.22, 1.13, -1.58],
[ 1.71, 0.64, -0.43, 0.34, 0.46]])
- Why are the values above different from the values in
stdscaler.transform(sales.iloc[:, 1:].tail(5))?
Interpreting standardized regression coefficients¶
- Now that we have a technique for standardizing the feature columns of
sales, let's fit a new regression object.
sales_model_std = LinearRegression()
sales_model_std.fit(X=stdscaler.transform(sales.iloc[:, 1:]),
y=sales.iloc[:, 0])
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
- Let's now look at the resulting coefficients, and compare them to the coefficients before we standardized.
pd.DataFrame().assign(
column=sales.columns[1:],
original_coef=sales_model.coef_,
standardized_coef=sales_model_std.coef_
).set_index('column')
| original_coef | standardized_coef | |
|---|---|---|
| column | ||
| sq_ft | 16.20 | 31.97 |
| inventory | 0.17 | 32.76 |
| advertising | 11.53 | 42.69 |
| district_size | 13.58 | 68.50 |
| competing_stores | -5.31 | -25.52 |
- Did the performance of the resulting model change?
mean_squared_error(sales.iloc[:, 0],
sales_model.predict(sales.iloc[:, 1:]))
242.27445717154964
mean_squared_error(sales.iloc[:, 0],
sales_model_std.predict(stdscaler.transform(sales.iloc[:, 1:])))
242.27445717154956
- No!
The span of the design matrix did not change, so the predictions did not change. It's just the coefficients that changed.
Key takeaways¶
- The result of standardizing each feature (separately!) is that the units of each feature are on the same scale.
- There's no need to standardize the outcome (
'net_sales'here), since it's not being compared to anything. - Also, we can't standardize the column of all 1s.
- There's no need to standardize the outcome (
- Then, solve the normal equations. The resulting $w_0^*, w_1^*, \ldots, w_d^*$ are called the standardized regression coefficients.
- Standardized regression coefficients can be directly compared to one another.
- As we saw on the previous slide, standardizing each feature does not change the MSE of the resulting hypothesis function!
StandardScaler summary¶
| Property | Example | Description |
|---|---|---|
| Initialize with parameters | stdscaler = StandardScaler() |
z-score the data (no parameters) |
| Fit the transformer | stdscaler.fit(X) |
Compute the mean and SD of X |
| Transform data in a dataset | feat = stdscaler.transform(X_new) |
z-score X_new with mean and SD of X |
| Fit and transform | stdscaler.fit_transform(X) |
Compute the mean and SD of X, then z-score X |
What's next?¶
- Even though we have a
OneHotEncodertransformer object, to actually use one hot encoding to make predictions, we need to:- Manually instantiate a
OneHotEncoderobject, and thenfitit. - Create a new design matrix by taking the result of calling
transformon theOneHotEncoderobject and concatenating other relevant numerical columns. - Manually instantiate a
LinearRegressionobject, and thenfitit using the result of the above step.
- Manually instantiate a
- As we build more and more sophisticated models, it will be challenging to keep track of all of these individual steps ourselves.
- As such, we often build Pipelines.
