from lec_utils import *
Announcements 📣¶
- Homework 1 is due tonight, though note that you have 6 slip days to use during the semester, and you can use up to 2 slip days on any homework (see here for policy details).
Post on Ed or come to Office Hours for help! We're using a queue for office hours now – access it from practicaldsc.org/calendar.
Homework 2 will be released tomorrow.
We'll make an Ed announcement anytime an assignment is released.In discussion tomorrow, we'll cover past exam problems on paper related to this week's material.
Check out the Resources tab on the course website, with links to lots of supplementary resources.
New link: EECS 201: Computer Science Pragmatics. Look here for help with Terminal commands,git, etc.
Agenda¶
- Randomness and simulation.
- Introduction to
pandasDataFrames.- Selecting columns from a DataFrame.
- Selecting rows from a DataFrame.
Remember to follow along in lecture by accessing the "blank" lecture notebook in our public GitHub repository.
Question 🤔 (Answer at practicaldsc.org/q)
Remember that you can always ask questions anonymously at the link above!When is your birthday?
Randomness and simulation¶
We'll start by exploring a useful application of numpy in the field of probability and statistics: simulation!
np.random¶
The submodule np.random contains various functions that produce random results.
These use pseudo-random number generators to generate random-seeming sequences of results.
# Run this cell multiple times!
# Returns a random integer between 1 and 6, inclusive.
np.random.randint(1, 7)
3
# Returns a random real number between 0 and 1.
np.random.random()
0.24976347964756174
# Returns a randomly selected element from the provided list, 5 times.
np.random.choice(['H', 'T'], 5)
array(['T', 'H', 'T', 'T', 'T'], dtype='<U1')
# Returns the number of occurrences of each outcome
# in 12 trials of an experiment in which
# outcome 1 happens 60% of the time and
# outcome 2 happens 40% of the time.
np.random.multinomial(12, [0.6, 0.4])
array([3, 9])
Simulations¶
- Often, we'll want to estimate the probability of an event, but it may not be possible – or we may not know how – to calculate the probability exactly.
e.g., the probability that I see between 40 and 50 heads when I flip a fair coin 100 times.
- Or, we may have a theoretical answer, and want to validate it using another approach.
In such cases, we can use the power of simulation. We can:
- Figure out how to simulate one run of the experiment.
e.g., figure out how to get Python to flip a fair coin 100 times and count the number of heads. - Repeat the experiment many, many times.
- Compute the fraction of experiments in which our event occurs, and use this fraction as an estimate of the probability of our event.
This is the basis of Monte Carlo Methods.
- Figure out how to simulate one run of the experiment.
- Theory tells us that the more repetitions we perform of our experiment, the closer our fraction will be to the true probability of the event!
Specifically, the Law of Large Numbers tells us this.
Example: Coin flipping¶
- Question: What is the probability that I see between 40 and 50 heads, inclusive, when I flip a fair coin 100 times?
- Step 1: Figure out how to simulate one run of the experiment.
e.g., figure out how to get Python to flip a fair coin 100 times and count the number of heads.
(np.random.choice(['H', 'T'], 100) == 'H').sum()
48
np.random.multinomial(100, [0.5, 0.5])[0]
55
def num_heads():
return np.random.multinomial(100, [0.5, 0.5])[0]
num_heads()
49
- Step 2: Repeat the experiment many, many times.
In other words, run the cell above lots of times and store the results somewhere.
outcomes = np.array([])
for _ in range(10_000):
# Note that with arrays, append is a FUNCTION,
# not a METHOD, and is NOT destructive,
# unlike with lists!
outcomes = np.append(outcomes, num_heads())
- Step 3: Compute the fraction of experiments in which our event occurs, and use this fraction as an estimate of the probability of our event.
px.histogram(outcomes)
((outcomes >= 40) & (outcomes <= 50)).mean()
0.5207
- This is remarkably close to the true, theoretical answer!
from scipy.stats import binom
binom.cdf(50, 100, 0.5) - binom.cdf(39, 100, 0.5)
0.5221945185847372
Question 🤔 (Answer at practicaldsc.org/q)
What questions do you have about our coin flipping simulation?
Can you think of a way to perform the same simulation without any for-loops?
Example: The Birthday Paradox¶
- There are ~80 students in the room right now. What are the chances at least 2 students share the same birthday?
- In general, how many people must be in a room such that there's a 50% chance that at least 2 students share the same birthday?
- Let's define a function,
estimated_probability, which takes in a class size,n, and returns the probability that in a class ofnstudents, at least 2 students share the same birthday.
def simulate_classroom(n):
# This helper function should take in a class size, n,
# and return True if a simulated classroom of size n
# has at least 2 students with the same birthday
# and False otherwise.
# This is not the most efficient solution, but works for now.
options = np.arange(1, 366)
chosen_options = np.random.choice(options, n, replace=True)
return len(chosen_options) != len(np.unique(chosen_options))
def estimated_probability(n):
return np.mean([simulate_classroom(n) for _ in range(10_000)])
estimated_probability(80)
0.9999
- With 80 students, it's almost certain that 2 share the same birthday!
What's the minimum class size we'd need for a 50% chance that at least 2 share the same birthday?
probs = [estimated_probability(n) for n in range(1, 51)]
(
px
.bar(x=range(1, 51),
y=probs,
title='Probability that at least 2 students share the<br>same birthday in a class of n students')
.update_xaxes(title='$n$')
.update_yaxes(title='Probability')
)
- Lower than you might think!
Introduction to pandas DataFrames¶
Let's finally start working with real datasets! 🎉
Note that we're going to cover a lot of code quickly. The point of lecture is to expose you to what's possible; you can look at the notebook later for the details.
pandas¶

pandasis the Python library for tabular data manipulation.
- Before
pandaswas developed, the standard data science workflow involved using multiple languages (Python, R, Java) in a single project.
- Wes McKinney, the original developer of
pandas, wanted a library which would allow everything to be done in Python.
Python is faster to develop in than Java or C++, and is more general-purpose than R.
Importing pandas and related libraries¶
pandas is almost always imported in conjunction with numpy.
import pandas as pd
import numpy as np
pandas data structures¶
There are three key data structures at the core of pandas.
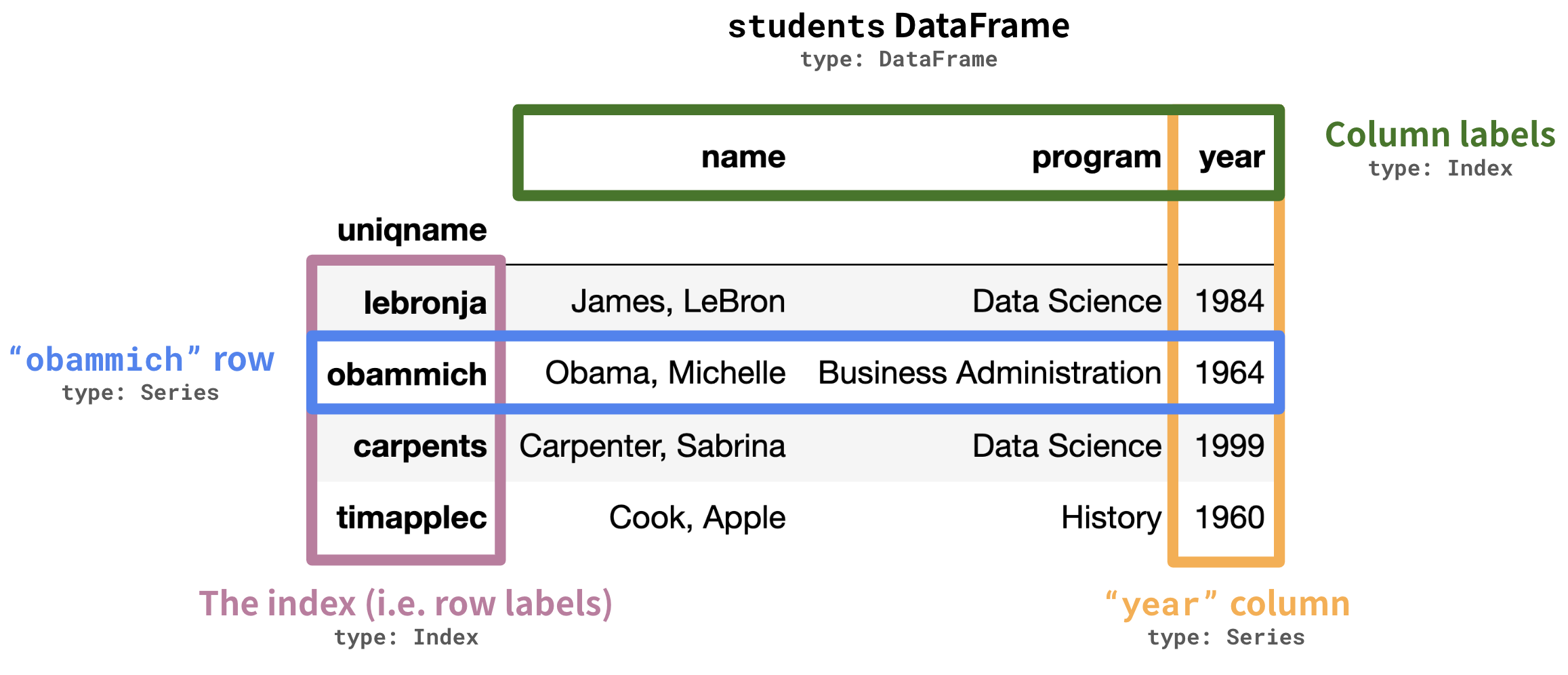 An example DataFrame.
An example DataFrame.
- DataFrame: 2 dimensional tables. These have rows and columns.
- Series: 1 dimensional array-like object, representing a row or column.
Like arrays, Series contain data of the same type. The plural of Series is also Series.
- Index: Sequence of row or column labels. When we say "the index", we're referring to the sequence of row labels.
The index –'lebronja','obammich','carpents', and'timapplec'in the example above – is not a column!
Column names –'name','program', and'year'in the example above – are stored as strings, and the sequence of column names is also an index.
Example: Dog Breeds 🐶¶
- The dataset we'll work comes from the American Kennel Club. Here's a cool plot made using our dataset.
- We'll usually work with data stored in the CSV format. CSV stands for "comma-separated values."
- We can read in a CSV using
pd.read_csv(path). The path should be relative to your notebook; if the file is in the same folder as your notebook, this is just the name of the file (as a string).
- Today's dataset is stored
'data/dogs43.csv'– open it up and see what it looks like!
# The "cat" shell command shows you the contents of a file.
!cat data/dogs43.csv
breed,kind,lifetime_cost,longevity,size,weight,height Brittany,sporting,22589.0,12.92,medium,35.0,19.0 Cairn Terrier,terrier,21992.0,13.84,small,14.0,10.0 English Cocker Spaniel,sporting,18993.0,11.66,medium,30.0,16.0 Cocker Spaniel,sporting,24330.0,12.5,small,25.0,14.5 Shetland Sheepdog,herding,21006.0,12.53,small,22.0,14.5 Siberian Husky,working,22049.0,12.58,medium,47.5,21.75 Lhasa Apso,non-sporting,22031.0,13.92,small,15.0,10.5 Miniature Schnauzer,terrier,20087.0,11.81,small,15.5,13.0 Chihuahua,toy,26250.0,16.5,small,5.5,5.0 English Springer Spaniel,sporting,21946.0,12.54,medium,45.0,19.5 German Shorthaired Pointer,sporting,25842.0,11.46,large,62.5,24.0 Pointer,sporting,24445.0,12.42,large,59.5,25.5 Tibetan Spaniel,non-sporting,25549.0,14.42,small,12.0,10.0 Labrador Retriever,sporting,21299.0,12.04,medium,67.5,23.0 Maltese,toy,19084.0,12.25,small,5.0,9.0 Shih Tzu,toy,21152.0,13.2,small,12.5,9.75 Irish Setter,sporting,20323.0,11.63,large,65.0,26.0 Golden Retriever,sporting,21447.0,12.04,medium,60.0,22.75 Chesapeake Bay Retriever,sporting,16697.0,9.48,large,67.5,23.5 Tibetan Terrier,non-sporting,20336.0,12.31,small,24.0,15.5 Gordon Setter,sporting,19605.0,11.1,large,62.5,25.0 Pug,toy,18527.0,11.0,medium,16.0,16.0 Norfolk Terrier,terrier,24308.0,13.07,small,12.0,9.5 English Toy Spaniel,toy,17521.0,10.1,small,11.0,10.0 Cavalier King Charles Spaniel,toy,18639.0,11.29,small,15.5,12.5 Basenji,hound,22096.0,13.58,medium,23.0,16.5 Staffordshire Bull Terrier,terrier,21650.0,12.05,medium,31.0,15.0 Pembroke Welsh Corgi,herding,23978.0,12.25,small,26.0,11.0 Clumber Spaniel,sporting,18084.0,10.0,medium,70.0,18.5 Dandie Dinmont Terrier,terrier,21633.0,12.17,small,21.0,9.0 Giant Schnauzer,working,26686.0,10.0,large,77.5,25.5 Scottish Terrier,terrier,17525.0,10.69,small,20.0,10.0 Kerry Blue Terrier,terrier,17240.0,9.4,medium,36.5,18.5 Afghan Hound,hound,24077.0,11.92,large,55.0,26.0 Newfoundland,working,19351.0,9.32,large,125.0,27.0 Rhodesian Ridgeback,hound,16530.0,9.1,large,77.5,25.5 Borzoi,hound,16176.0,9.08,large,82.5,28.0 Bull Terrier,terrier,18490.0,10.21,medium,60.0,21.5 Alaskan Malamute,working,21986.0,10.67,large,80.0,24.0 Bloodhound,hound,13824.0,6.75,large,85.0,25.0 Bullmastiff,working,13936.0,7.57,large,115.0,25.5 Mastiff,working,13581.0,6.5,large,175.0,30.0 Saint Bernard,working,20022.0,7.78,large,155.0,26.5
dogs = pd.read_csv('data/dogs43.csv')
dogs
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 40 | Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 7 columns
Exploring our first DataFrame¶
- To extract the first or last few rows of a DataFrame, use the
headortailmethods.
Like most DataFrame methods,headandtaildon't modify the original DataFrame!
dogs.head(3)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
dogs.tail(2)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
dogs
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 40 | Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 7 columns
- The
shapeattribute returns the DataFrame's number of rows and columns.
Sure, we can see 👀 that it says 43 rows x 7 columns above, but theshapeattribute allows us to write code involving the number of rows/columns.
# Note that the index – 0, 1, 2, ... – does **not** count as a column!
dogs.shape
(43, 7)
Sorting¶
- To sort by a column, use the
sort_valuesmethod.ascending=Falseis a keyword argument, meaning you need to specify the name of the argument to use it. You've seen some examples of this in theplotlypart of Homework 1.
# Note that the index is no longer 0, 1, 2, ...!
dogs.sort_values('height', ascending=False)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 36 | Borzoi | hound | 16176.0 | 9.08 | large | 82.5 | 28.0 |
| 34 | Newfoundland | working | 19351.0 | 9.32 | large | 125.0 | 27.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 29 | Dandie Dinmont Terrier | terrier | 21633.0 | 12.17 | small | 21.0 | 9.0 |
| 14 | Maltese | toy | 19084.0 | 12.25 | small | 5.0 | 9.0 |
| 8 | Chihuahua | toy | 26250.0 | 16.50 | small | 5.5 | 5.0 |
43 rows × 7 columns
- We can also sort by multiple columns!
This sorts by'height', then breaks ties by'longevity'. Note the difference in the last three rows between this DataFrame and the one above.
dogs.sort_values(['height', 'longevity'], ascending=False)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 36 | Borzoi | hound | 16176.0 | 9.08 | large | 82.5 | 28.0 |
| 34 | Newfoundland | working | 19351.0 | 9.32 | large | 125.0 | 27.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 14 | Maltese | toy | 19084.0 | 12.25 | small | 5.0 | 9.0 |
| 29 | Dandie Dinmont Terrier | terrier | 21633.0 | 12.17 | small | 21.0 | 9.0 |
| 8 | Chihuahua | toy | 26250.0 | 16.50 | small | 5.5 | 5.0 |
43 rows × 7 columns
Setting the index¶
- Think of each row's index as its unique identifier or name. The default index when we create a DataFrame using
pd.read_csvis 0, 1, 2, 3, ...
Think of the index of a DataFrame like a "key" in a dictionary (Python) or map (C++).
dogs
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 40 | Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 7 columns
dogs.index
RangeIndex(start=0, stop=43, step=1)
- Often, we like to set the index of a DataFrame to a unique identifier if we have one available. We can do so with the
set_indexmethod.
We'll see the real benefit of this shortly.
dogs.set_index('breed')
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
# The above cell didn't involve an assignment statement, so dogs was unchanged.
dogs
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 40 | Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 7 columns
# By reassigning dogs, our changes will persist.
# Note that we can't run this cell twice! Try it and see what happens.
dogs = dogs.set_index('breed')
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
# There used to be 7 columns, but now there are only 6!
# The index is **not** a column!
dogs.shape
(43, 6)
dogs.index
Index(['Brittany', 'Cairn Terrier', 'English Cocker Spaniel', 'Cocker Spaniel',
'Shetland Sheepdog', 'Siberian Husky', 'Lhasa Apso',
'Miniature Schnauzer', 'Chihuahua', 'English Springer Spaniel',
'German Shorthaired Pointer', 'Pointer', 'Tibetan Spaniel',
'Labrador Retriever', 'Maltese', 'Shih Tzu', 'Irish Setter',
'Golden Retriever', 'Chesapeake Bay Retriever', 'Tibetan Terrier',
'Gordon Setter', 'Pug', 'Norfolk Terrier', 'English Toy Spaniel',
'Cavalier King Charles Spaniel', 'Basenji',
'Staffordshire Bull Terrier', 'Pembroke Welsh Corgi', 'Clumber Spaniel',
'Dandie Dinmont Terrier', 'Giant Schnauzer', 'Scottish Terrier',
'Kerry Blue Terrier', 'Afghan Hound', 'Newfoundland',
'Rhodesian Ridgeback', 'Borzoi', 'Bull Terrier', 'Alaskan Malamute',
'Bloodhound', 'Bullmastiff', 'Mastiff', 'Saint Bernard'],
dtype='object', name='breed')
Activity
Assign tallest_breed to the name, as a string, of the tallest breed in the dataset. Answer using pandas code, i.e. don't look at the dataset and hard-code the answer.
tallest_breed = dogs.sort_values('height', ascending=False).index[0]
tallest_breed
'Mastiff'
Question 🤔 (Answer at practicaldsc.org/q)
What are your thoughts on the activities in lecture?
- A. I really hate them, get rid of them and spend more time lecturing.
- B. I don't mind them.
- C. I'm neutral – I wouldn't be sad to see them go, but don't mind if they're there.
- D. I like them.
- E. I really love them, do more!
💡 Pro-Tip: Displaying more rows/columns¶
Sometimes, you just want pandas to display a lot of rows and columns. You can use this helper function to do that.
from IPython.display import display
def display_df(df, rows=pd.options.display.max_rows, cols=pd.options.display.max_columns):
"""Displays n rows and cols from df."""
with pd.option_context("display.max_rows", rows,
"display.max_columns", cols):
display(df)
display_df(dogs.sort_values('weight', ascending=False), rows=43)
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.00 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.50 |
| Newfoundland | working | 19351.0 | 9.32 | large | 125.0 | 27.00 |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.50 |
| Bloodhound | hound | 13824.0 | 6.75 | large | 85.0 | 25.00 |
| Borzoi | hound | 16176.0 | 9.08 | large | 82.5 | 28.00 |
| Alaskan Malamute | working | 21986.0 | 10.67 | large | 80.0 | 24.00 |
| Rhodesian Ridgeback | hound | 16530.0 | 9.10 | large | 77.5 | 25.50 |
| Giant Schnauzer | working | 26686.0 | 10.00 | large | 77.5 | 25.50 |
| Clumber Spaniel | sporting | 18084.0 | 10.00 | medium | 70.0 | 18.50 |
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium | 67.5 | 23.00 |
| Chesapeake Bay Retriever | sporting | 16697.0 | 9.48 | large | 67.5 | 23.50 |
| Irish Setter | sporting | 20323.0 | 11.63 | large | 65.0 | 26.00 |
| German Shorthaired Pointer | sporting | 25842.0 | 11.46 | large | 62.5 | 24.00 |
| Gordon Setter | sporting | 19605.0 | 11.10 | large | 62.5 | 25.00 |
| Bull Terrier | terrier | 18490.0 | 10.21 | medium | 60.0 | 21.50 |
| Golden Retriever | sporting | 21447.0 | 12.04 | medium | 60.0 | 22.75 |
| Pointer | sporting | 24445.0 | 12.42 | large | 59.5 | 25.50 |
| Afghan Hound | hound | 24077.0 | 11.92 | large | 55.0 | 26.00 |
| Siberian Husky | working | 22049.0 | 12.58 | medium | 47.5 | 21.75 |
| English Springer Spaniel | sporting | 21946.0 | 12.54 | medium | 45.0 | 19.50 |
| Kerry Blue Terrier | terrier | 17240.0 | 9.40 | medium | 36.5 | 18.50 |
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.00 |
| Staffordshire Bull Terrier | terrier | 21650.0 | 12.05 | medium | 31.0 | 15.00 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.00 |
| Pembroke Welsh Corgi | herding | 23978.0 | 12.25 | small | 26.0 | 11.00 |
| Cocker Spaniel | sporting | 24330.0 | 12.50 | small | 25.0 | 14.50 |
| Tibetan Terrier | non-sporting | 20336.0 | 12.31 | small | 24.0 | 15.50 |
| Basenji | hound | 22096.0 | 13.58 | medium | 23.0 | 16.50 |
| Shetland Sheepdog | herding | 21006.0 | 12.53 | small | 22.0 | 14.50 |
| Dandie Dinmont Terrier | terrier | 21633.0 | 12.17 | small | 21.0 | 9.00 |
| Scottish Terrier | terrier | 17525.0 | 10.69 | small | 20.0 | 10.00 |
| Pug | toy | 18527.0 | 11.00 | medium | 16.0 | 16.00 |
| Miniature Schnauzer | terrier | 20087.0 | 11.81 | small | 15.5 | 13.00 |
| Cavalier King Charles Spaniel | toy | 18639.0 | 11.29 | small | 15.5 | 12.50 |
| Lhasa Apso | non-sporting | 22031.0 | 13.92 | small | 15.0 | 10.50 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.00 |
| Shih Tzu | toy | 21152.0 | 13.20 | small | 12.5 | 9.75 |
| Tibetan Spaniel | non-sporting | 25549.0 | 14.42 | small | 12.0 | 10.00 |
| Norfolk Terrier | terrier | 24308.0 | 13.07 | small | 12.0 | 9.50 |
| English Toy Spaniel | toy | 17521.0 | 10.10 | small | 11.0 | 10.00 |
| Chihuahua | toy | 26250.0 | 16.50 | small | 5.5 | 5.00 |
| Maltese | toy | 19084.0 | 12.25 | small | 5.0 | 9.00 |
Selecting columns from a DataFrame¶
In order to answer questions involving our data, we'll need to be able to access the values stored in individual columns.
Selecting columns with []¶
- The most common way to select a subset of the columns in a DataFrame is by using the
[]operator.
This is just like when we accessed values in a dictionary based on their key.
- Specifying a column name returns the column as a Series.
- Specifying a list of column names returns a DataFrame.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
# Returns a Series.
dogs['kind']
breed
Brittany sporting
Cairn Terrier terrier
English Cocker Spaniel sporting
...
Bullmastiff working
Mastiff working
Saint Bernard working
Name: kind, Length: 43, dtype: object
# Returns a DataFrame.
dogs[['kind', 'size']]
| kind | size | |
|---|---|---|
| breed | ||
| Brittany | sporting | medium |
| Cairn Terrier | terrier | small |
| English Cocker Spaniel | sporting | medium |
| ... | ... | ... |
| Bullmastiff | working | large |
| Mastiff | working | large |
| Saint Bernard | working | large |
43 rows × 2 columns
# 🤔
dogs[['kind']]
| kind | |
|---|---|
| breed | |
| Brittany | sporting |
| Cairn Terrier | terrier |
| English Cocker Spaniel | sporting |
| ... | ... |
| Bullmastiff | working |
| Mastiff | working |
| Saint Bernard | working |
43 rows × 1 columns
- As an aside: when you get an error message in Python, the most informative part is usually at the bottom!
So, if you're posting about your error on Ed, or debugging with us in office hours, show us the bottom first.
# Breeds are stored in the index, which is not a column!
dogs['breed']
--------------------------------------------------------------------------- KeyError Traceback (most recent call last) File ~/miniforge3/envs/pds/lib/python3.10/site-packages/pandas/core/indexes/base.py:3790, in Index.get_loc(self, key) 3789 try: -> 3790 return self._engine.get_loc(casted_key) 3791 except KeyError as err: File index.pyx:152, in pandas._libs.index.IndexEngine.get_loc() File index.pyx:181, in pandas._libs.index.IndexEngine.get_loc() File pandas/_libs/hashtable_class_helper.pxi:7080, in pandas._libs.hashtable.PyObjectHashTable.get_item() File pandas/_libs/hashtable_class_helper.pxi:7088, in pandas._libs.hashtable.PyObjectHashTable.get_item() KeyError: 'breed' The above exception was the direct cause of the following exception: KeyError Traceback (most recent call last) Cell In[40], line 2 1 # Breeds are stored in the index, which is not a column! ----> 2 dogs['breed'] File ~/miniforge3/envs/pds/lib/python3.10/site-packages/pandas/core/frame.py:3896, in DataFrame.__getitem__(self, key) 3894 if self.columns.nlevels > 1: 3895 return self._getitem_multilevel(key) -> 3896 indexer = self.columns.get_loc(key) 3897 if is_integer(indexer): 3898 indexer = [indexer] File ~/miniforge3/envs/pds/lib/python3.10/site-packages/pandas/core/indexes/base.py:3797, in Index.get_loc(self, key) 3792 if isinstance(casted_key, slice) or ( 3793 isinstance(casted_key, abc.Iterable) 3794 and any(isinstance(x, slice) for x in casted_key) 3795 ): 3796 raise InvalidIndexError(key) -> 3797 raise KeyError(key) from err 3798 except TypeError: 3799 # If we have a listlike key, _check_indexing_error will raise 3800 # InvalidIndexError. Otherwise we fall through and re-raise 3801 # the TypeError. 3802 self._check_indexing_error(key) KeyError: 'breed'
dogs.index
Index(['Brittany', 'Cairn Terrier', 'English Cocker Spaniel', 'Cocker Spaniel',
'Shetland Sheepdog', 'Siberian Husky', 'Lhasa Apso',
'Miniature Schnauzer', 'Chihuahua', 'English Springer Spaniel',
'German Shorthaired Pointer', 'Pointer', 'Tibetan Spaniel',
'Labrador Retriever', 'Maltese', 'Shih Tzu', 'Irish Setter',
'Golden Retriever', 'Chesapeake Bay Retriever', 'Tibetan Terrier',
'Gordon Setter', 'Pug', 'Norfolk Terrier', 'English Toy Spaniel',
'Cavalier King Charles Spaniel', 'Basenji',
'Staffordshire Bull Terrier', 'Pembroke Welsh Corgi', 'Clumber Spaniel',
'Dandie Dinmont Terrier', 'Giant Schnauzer', 'Scottish Terrier',
'Kerry Blue Terrier', 'Afghan Hound', 'Newfoundland',
'Rhodesian Ridgeback', 'Borzoi', 'Bull Terrier', 'Alaskan Malamute',
'Bloodhound', 'Bullmastiff', 'Mastiff', 'Saint Bernard'],
dtype='object', name='breed')
Useful Series methods¶
- A Series is like an array, but with an index.
- There are a variety of useful methods that work on Series. You can see the entire list here. Many methods that work on a Series will also work on DataFrames, as we'll soon see.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
# What are the unique kinds of dogs?
dogs['kind'].unique()
array(['sporting', 'terrier', 'herding', 'working', 'non-sporting', 'toy',
'hound'], dtype=object)
# How many unique kinds of dogs are there?
dogs['kind'].nunique()
7
# What's the distribution of kinds?
# value_counts is super useful – and I love asking exam questions about it!
dogs['kind'].value_counts()
kind sporting 12 terrier 8 working 7 toy 6 hound 5 non-sporting 3 herding 2 Name: count, dtype: int64
# What's the mean of the 'longevity' column?
dogs['longevity'].mean()
11.340697674418605
# Tell me more about the 'weight' column.
dogs['weight'].describe()
count 43.00
mean 49.35
std 39.42
...
50% 36.50
75% 67.50
max 175.00
Name: weight, Length: 8, dtype: float64
# Sort the 'lifetime_cost' column. Note that here we're using sort_values on a Series, not a DataFrame!
dogs['lifetime_cost'].sort_values()
breed
Mastiff 13581.0
Bloodhound 13824.0
Bullmastiff 13936.0
...
German Shorthaired Pointer 25842.0
Chihuahua 26250.0
Giant Schnauzer 26686.0
Name: lifetime_cost, Length: 43, dtype: float64
# Gives us the index of the largest value, not the largest value itself.
# Note that this makes our Activity from a few slides ago way easier!
dogs['height'].idxmax()
'Mastiff'
Activity
Complete the implementation of the function average_heaviest, which takes in a positive integer n and returns the mean 'lifetime_cost' of the top n heaviest breeds. Example behavior is given below.
>>> average_heaviest(5)
16142.8
>>> average_heaviest(1)
13581.0
We won't have time to try this activity in lecture, but the answer is posted in lec04-filled.ipynb and in the "filled html" link on the course website.
def average_heaviest(n):
return (
dogs
.sort_values('weight', ascending=False)
.head(n)
['lifetime_cost']
.mean()
)
average_heaviest(5)
16142.8
average_heaviest(1)
13581.0
Series support vectorized operations¶
- Series operations are vectorized, just like with arrays.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
- Example: If I adopt a puppy next year, when should I expect them to live until?
2025 + dogs['longevity']
breed
Brittany 2037.92
Cairn Terrier 2038.84
English Cocker Spaniel 2036.66
...
Bullmastiff 2032.57
Mastiff 2031.50
Saint Bernard 2032.78
Name: longevity, Length: 43, dtype: float64
- Example: What is the average cost per year to maintain each breed?
dogs['lifetime_cost'] / dogs['longevity']
breed
Brittany 1748.37
Cairn Terrier 1589.02
English Cocker Spaniel 1628.90
...
Bullmastiff 1840.95
Mastiff 2089.38
Saint Bernard 2573.52
Length: 43, dtype: float64
- Example: Which breed is the cheapest to own per year, on average? The most expensive?
(dogs['lifetime_cost'] / dogs['longevity']).sort_values()
breed
Maltese 1557.88
Lhasa Apso 1582.69
Cairn Terrier 1589.02
...
German Shorthaired Pointer 2254.97
Saint Bernard 2573.52
Giant Schnauzer 2668.60
Length: 43, dtype: float64
(dogs['lifetime_cost'] / dogs['longevity']).idxmax()
'Giant Schnauzer'
Activity
Assign bmis to a Series containing the Body Mass Index (BMI) of each breed, using the following formula:
Note that in dogs, weights are measured in pounds and heights are measured in inches. Use the following conversion factors:
1 inch = 2.54 cm = 0.0254 m
Your solution can span multiple lines, and you can define intermediate variables if you need to (we did).
weight_kg = dogs['weight'] / 2.2
height_m = dogs['height'] * 2.54 / 100
bmis = weight_kg / (height_m ** 2)
bmis
breed
Brittany 68.31
Cairn Terrier 98.64
English Cocker Spaniel 82.56
...
Bullmastiff 124.60
Mastiff 137.00
Saint Bernard 155.51
Length: 43, dtype: float64
Aside: Visualization¶
- We'll spend more time talking about when to create which types of visualizations in a few lectures.
- But for now, you can start exploring how the DataFrame
plotmethod works!
dogs.plot(kind='scatter', x='weight', y='longevity')
# Hover over a point and see what happens!
(
dogs
.reset_index()
.plot(kind='scatter', x='weight', y='longevity', color='size', hover_name='breed',
title='Longevity vs. Weight for 43 Dog Breeds')
)
(
dogs['kind']
.value_counts()
.sort_values()
.plot(kind='barh', title='Distribution of Dog Kinds')
)
Selecting slices of a DataFrame¶
Now that we know how to access specific columns in a dataset, how do we access specific rows? Or even individual values?
Use loc to slice rows and columns using labels¶
locstands for "location".
- The
locindexer works similarly to slicing 2D arrays, but it uses row labels and column labels, not positions.
Remember, the "index" refers to the row labels.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
# The first argument is the row label, i.e. the index value.
# ↓
dogs.loc['Pug', 'longevity']
# ↑
# The second argument is the column label.
11.0
- As an aside,
locis not a method – it's an indexer.
type(dogs.loc)
pandas.core.indexing._LocIndexer
type(dogs.sort_values)
method
loc is flexible 🧘¶
You can provide a sequence (list, array, Series) as either argument to loc.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
dogs.loc[['Cocker Spaniel', 'Labrador Retriever'], 'size']
breed Cocker Spaniel small Labrador Retriever medium Name: size, dtype: object
dogs.loc[['Cocker Spaniel', 'Labrador Retriever'], ['kind', 'size', 'height']]
| kind | size | height | |
|---|---|---|---|
| breed | |||
| Cocker Spaniel | sporting | small | 14.5 |
| Labrador Retriever | sporting | medium | 23.0 |
# Note that the 'weight' column is included!
# loc, per the pandas documentation, is inclusive of both slicer endpoints.
dogs.loc[['Cocker Spaniel', 'Labrador Retriever'], 'lifetime_cost': 'weight']
| lifetime_cost | longevity | size | weight | |
|---|---|---|---|---|
| breed | ||||
| Cocker Spaniel | 24330.0 | 12.50 | small | 25.0 |
| Labrador Retriever | 21299.0 | 12.04 | medium | 67.5 |
dogs.loc[['Cocker Spaniel', 'Labrador Retriever'], :]
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Cocker Spaniel | sporting | 24330.0 | 12.50 | small | 25.0 | 14.5 |
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium | 67.5 | 23.0 |
# Shortcut for the line above.
dogs.loc[['Cocker Spaniel', 'Labrador Retriever']]
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Cocker Spaniel | sporting | 24330.0 | 12.50 | small | 25.0 | 14.5 |
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium | 67.5 | 23.0 |
Use iloc to slice rows and columns using positions¶
ilocstands for "integer location."
ilocis likeloc, but it selects rows and columns based off of integer positions only, just like with 2D arrays.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
# Try removing the iloc and see what happens!
dogs.iloc[1:15, :-2]
| kind | lifetime_cost | longevity | size | |
|---|---|---|---|---|
| breed | ||||
| Cairn Terrier | terrier | 21992.0 | 13.84 | small |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium |
| Cocker Spaniel | sporting | 24330.0 | 12.50 | small |
| ... | ... | ... | ... | ... |
| Tibetan Spaniel | non-sporting | 25549.0 | 14.42 | small |
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium |
| Maltese | toy | 19084.0 | 12.25 | small |
14 rows × 4 columns
ilocis often most useful when we sort first. For instance, to find the weight of the longest-living breed in the dataset:
dogs.sort_values('longevity', ascending=False)['weight'].iloc[0]
5.5
# Finding the breed itself involves sorting, but not iloc, since breeds are stored in the index.
dogs.sort_values('longevity', ascending=False).index[0]
'Chihuahua'
Activity
Among just the following four breeds, what is the height of the second tallest breed?
- Cocker Spaniel.
- Labrador Retriever.
- Irish Setter.
- Newfoundland.
Assign your answer to second_tallest_height below. Answer using pandas code, i.e. don't look at the dataset and hard-code the answer.
We won't have time to try this activity in lecture, but the answer is posted in lec04-filled.ipynb and in the "filled html" link on the course website.
second_tallest_height = (
dogs
.loc[['Cocker Spaniel', 'Labrador Retriever', 'Newfoundland', 'Irish Setter'], 'height']
.sort_values(ascending=False)
.iloc[1]
)
Querying¶
Okay, but what if we don't know anything about the position or index of a row we're looking for? How do we find rows that satisfy certain conditions?
Reflection¶
- So far, all of the questions we've been able to answer involved all of the rows in the dataset.
What's the weight of the longest-living breed? What's the average lifetime cost of all breeds? Which breed is third heaviest?
- We don't yet have a mechanism to answer questions about a specific subset of the dataset.
How many terriers are there? What's the average longevity of medium-sized breeds?
Querying¶
- Querying is the act selecting rows in a DataFrame that satisfy certain condition(s).
We sometimes call this "filtering."
- As we saw in Lecture 3, comparisons with arrays result in Boolean arrays. The same is true for Series – make a comparison with a Series, and the result is a Boolean Series!
- We can use comparisons along with the
locoperator to select specific rows from a DataFrame.
dogs
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 6 columns
dogs['kind'] == 'terrier'
breed
Brittany False
Cairn Terrier True
English Cocker Spaniel False
...
Bullmastiff False
Mastiff False
Saint Bernard False
Name: kind, Length: 43, dtype: bool
dogs.loc[dogs['kind'] == 'terrier']
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| Miniature Schnauzer | terrier | 20087.0 | 11.81 | small | 15.5 | 13.0 |
| Norfolk Terrier | terrier | 24308.0 | 13.07 | small | 12.0 | 9.5 |
| ... | ... | ... | ... | ... | ... | ... |
| Scottish Terrier | terrier | 17525.0 | 10.69 | small | 20.0 | 10.0 |
| Kerry Blue Terrier | terrier | 17240.0 | 9.40 | medium | 36.5 | 18.5 |
| Bull Terrier | terrier | 18490.0 | 10.21 | medium | 60.0 | 21.5 |
8 rows × 6 columns
# This gives us the number of terriers in the dataset.
dogs.loc[dogs['kind'] == 'terrier'].shape[0]
8
dogs.loc[dogs['weight'] >= 50]
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| German Shorthaired Pointer | sporting | 25842.0 | 11.46 | large | 62.5 | 24.0 |
| Pointer | sporting | 24445.0 | 12.42 | large | 59.5 | 25.5 |
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium | 67.5 | 23.0 |
| ... | ... | ... | ... | ... | ... | ... |
| Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
19 rows × 6 columns
# .str.contains is very useful!
dogs.loc[dogs.index.str.contains('Retriever')]
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium | 67.5 | 23.00 |
| Golden Retriever | sporting | 21447.0 | 12.04 | medium | 60.0 | 22.75 |
| Chesapeake Bay Retriever | sporting | 16697.0 | 9.48 | large | 67.5 | 23.50 |
# Because querying is so common, there's a shortcut:
dogs[dogs.index.str.contains('Retriever')]
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed | ||||||
| Labrador Retriever | sporting | 21299.0 | 12.04 | medium | 67.5 | 23.00 |
| Golden Retriever | sporting | 21447.0 | 12.04 | medium | 60.0 | 22.75 |
| Chesapeake Bay Retriever | sporting | 16697.0 | 9.48 | large | 67.5 | 23.50 |
# Empty DataFrame – not an error!
dogs.loc[dogs['kind'] == 'beaver']
| kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|
| breed |
Activity
Assign second_tallest to the 'size' of the second-tallest 'sporting' breed, as a string. Answer using pandas code, i.e. don't look at the dataset and hard-code the answer.
second_tallest = (
dogs[dogs['kind'] == 'sporting']
.sort_values('height', ascending=False)
['size']
.iloc[1]
)
second_tallest
'large'
Lingering questions¶
- How do we use multiple conditions?
We actually covered the answer to this in Lecture 3 – try it out yourself!
- There's a DataFrame
querymethod – how does it work, and how is it different from what we've seen here?
- How do we find the average longevity of every
'kind'without having to copy-paste a lot of code?
- We'll find out on Tuesday!